Figure 3.
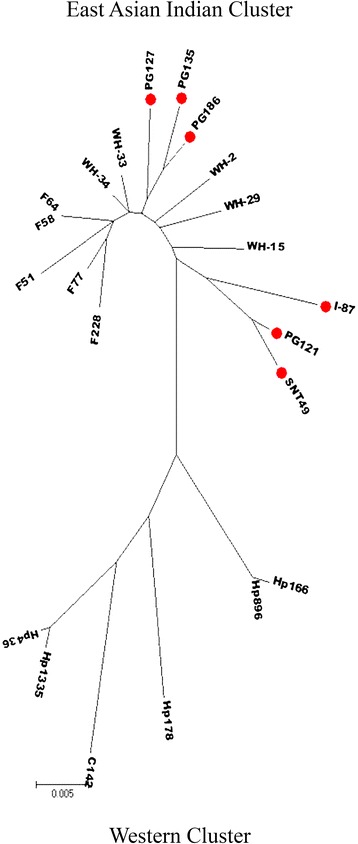
Phylogenetic tree constructed based on 1.8 kb segment of dupA gene of H. pylori (determined in this study and reported by others). This Maximum Likelihood tree was generated using Tamura 3 parameter model in MEGA6 software (version 6.0.5, AZ, USA). Representative strains from India are marked in Red dots (●, red circle). Sequences of non Indian strains were used here from public database. Indian and East Asian strains formed one cluster called group I and Brazilian and Colombian strains formed another cluster called group II. The length of the vertical bar indicates the number of nucleotide substitution per site. Bootstrap values of ≥ 70 are indicated at the nodes. H. pylori strain designations indicate the geographic origins, as follows: F, Japan; WH, China; PG or I or SNT, India; HP, Brazil; C, Colombia. GenBank: accession no. for the strains used in the study are given in the parentheses: F228 [GenBank: AB617836.1], F77 [GenBank: AB617834.1], F58 [GenBank: AB617835.1], F64 [GenBank: AB617833.1], WH-34 [GenBank: KC707844.1], WH-33 [GenBank: KC707843.1], F51 [GenBank: AB617832.1], PG127 [GenBank: Submission in process], PG135-1a [GenBank: JN379048.1], PG186 [GenBank: KC894690.1], WH-2 [GenBank: KC707837.1], WH-29 [GenBank: KC707842.1], WH-15 ([GenBank: KC707839.1], I-87 [GenBank: KC894692.1], I-121 [GenBank: KC894689.1], SNT49 GenBank: CP002983.1], Hp166.03 [GenBank: HM770857.1], Hp896.95 [GenBank: HM770862.1], Hp178.02 [GenBank: HQ228198.1], C142 [GenBank: AB196363.1], Hp1335.95 [GenBank: HQ228195.1], Hp436.95 [GenBank: HQ228197.1].
