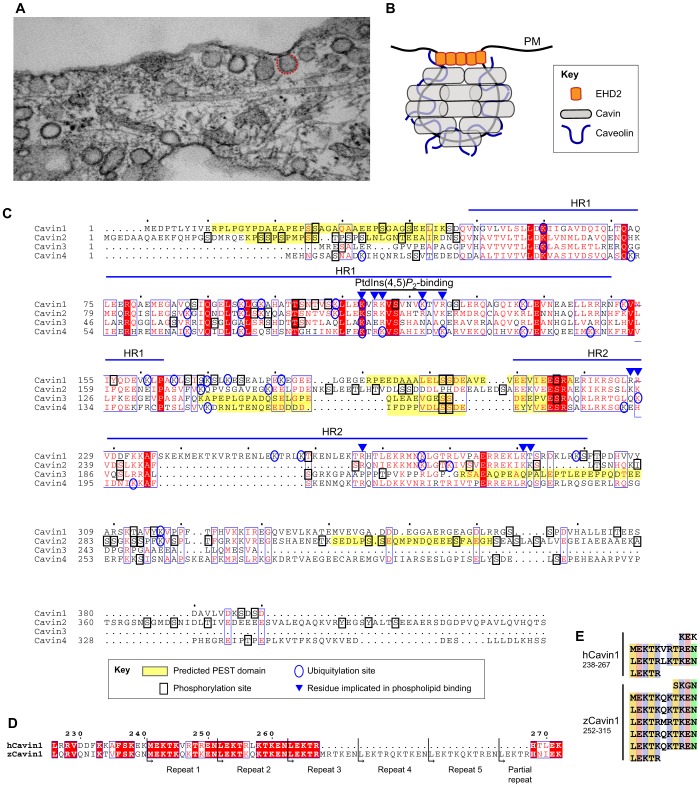
Fig. 1.
Caveolae and the cavin family of proteins. (A) Electron micrograph of caveolar bulbs (see caveolae marked by red dotted line) at the cell surface of a fibroblast cell. (B) Simplified model of the caveolar coat. Cavins are peripheral membrane proteins that coat the caveolar surface, with caveolins embedded in the interior membrane layer. EHD2 is known to associate with the caveolar neck and can modulate the dynamics of caveolae. (C) Secondary-structure-based sequence alignment of human cavins. Indicated on the alignment are predicted PEST domains (predicted by sPESTfind), and identified phosphorylation and ubiquitylation sites from the Phosphosite database (Hornbeck et al., 2012), as well as previously identified residues implicated in phospholipid interactions (Hansen et al., 2009; Kovtun et al., 2014). Amino acid residues in red are strictly conserved in the four cavins; amino acid residues surrounded by a blue box are highly conserved; amino acid residues highlighted red are conserved in residue type. (D) Putative 11-residue (hendecad/undecad) repeats unique to the HR2 domain of Cavin1, which – we propose – might form a coiled-coil structure. Portions of the Cavin1 sequences are shown highlighting the two repeats in human (h) Cavin1, and the five in zebrafish (z) Cavin1. (E) Alignment of the repeat elements shown in Fig. 1D; colour-coding indicates conserved hydrophobic (yellow), acidic (orange), basic (blue) and polar (green) residues.