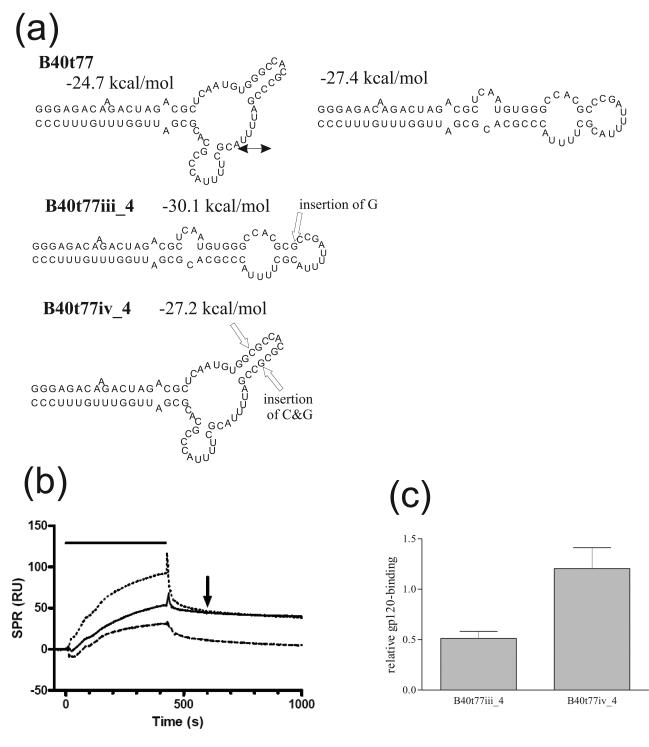
Figure 1. Confirmation of the branched structure of functional aptamer B40t77.
(a) Structures of aptamer B40t77, which is predicted to fold into both the linear or branched conformations, and its mutant derivatives designed to adopt solely the linear (B40t77iii_4) or branched (B40t77iv_4) conformation. The positions of nucleotide insertions are shown by open arrows, together with the predicted free energy of each structure and double-headed arrows indicating conformational interconversion. Structures and free energies were predicted using mfold. Unless otherwise stated, in this figure and elsewhere, all purine nucleotides (G, A) have 2′-OH, and all pyrimidine nucleotides (U, C) are 2′-F modified. (b) Representative BIAcore sensorgram overlay showing binding of aptamers B40t77 (solid line), B40t77iii_4 (dashed line) and B40t77iv_4 (dotted line) to gp120. In this and subsequent figures binding responses were normalized by subtracting the response from a blank flow cell, the duration of the injection is indicated by the horizontal bar and the 600 s time-point is indicated by an arrow. (c) Aptamer binding responses to gp120 obtained in (b). Here and subsequently responses observed 600 seconds after the start of injection are expressed relative to the binding of parental aptamer B40t77 following subtraction of the response in the blank flow cell, and normalized for the molecular mass of each aptamer. Error bars represent the standard deviation of three replicates.