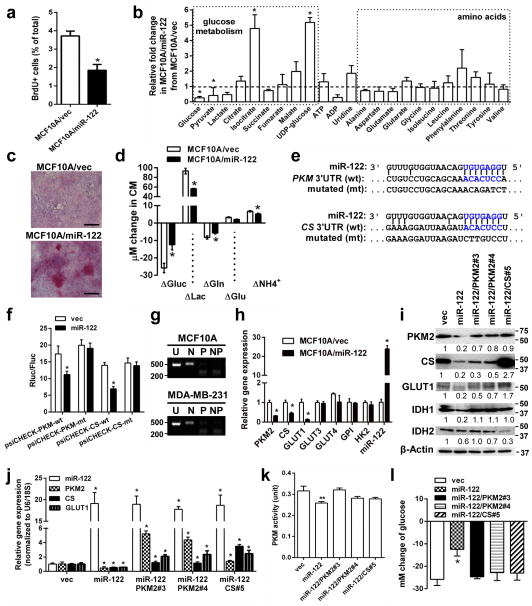
Figure 2.
MiR-122 suppresses glucose metabolism by downregulating PKM. (a) BrdU uptake in indicated cells were analysed by flow cytometry (n = 6 biological replicates). (b) Quantification of intracellular metabolites by NMR spectroscopy (n = 3 biological replicates). (c) Glycogen staining (red) in MCF10A/vec and MCF10A/miR-122 cells. Bar equals 100 μm. (d) Change of metabolites in the media after 72 h of culture (n = 6 biological replicates). (e) The predicted miR-122 binding site in the 3′UTR of the human PKM and CS gene. The corresponding sequence in the mutated (mt) version is also shown. (f) The psiCHECK reporters containing 3′UTR of human PKM and CS gene with wild-type (wt) or mutated (mt) miR-122 binding site were used to transfect MCF10A cells stably expressing miR-122 or the empty vector (as control). Luciferase activity was analysed at 48 h post-transfection (n = 6 extracts). (g) Determination of PKM isoforms expressed in MCF10A and MDA-MB-231. RNA was subjected to RT-PCR followed by digestion with NcoI (N), PstI (P), or both enzymes (NP), plus an uncut control (U). Products were separated on an agarose gel with Sybr safe. The presence of a PstI digestion site indicates the splicing isoform M2 whereas the NcoI site indicates isoform M1. Size of markers (in bp) are indicated. (h) RT-qPCR analysis showing the relative expression of indicated genes in MCF10A/miR-122 and MCF10A/vec cells (n = 6 extracts). (i) Western blot analysis in MCF10A/miR-122 and MCF10A/vec cells with restored expression of PKM2 and CS. Size of markers (in kDa) are indicated. (j) RT-qPCR analysis in selected colonies with restored expression of PKM2 and CS (n = 6 extracts). (k) PKM activity (unit) in 5 μg proteins in indicated cells (n = 6 extracts). (l) Change of glucose in the media after 72 h of culture of selected clones normalized to cell number (n = 3 biological replicates). * p < 0.05, ** p < 0.01 for all panels derived from Kruskal-Wallis test. Data are represented as mean ± SD in all panels except (c, e, g, i). Uncropped images of blots and gels are shown in Supplementary Fig. 5.