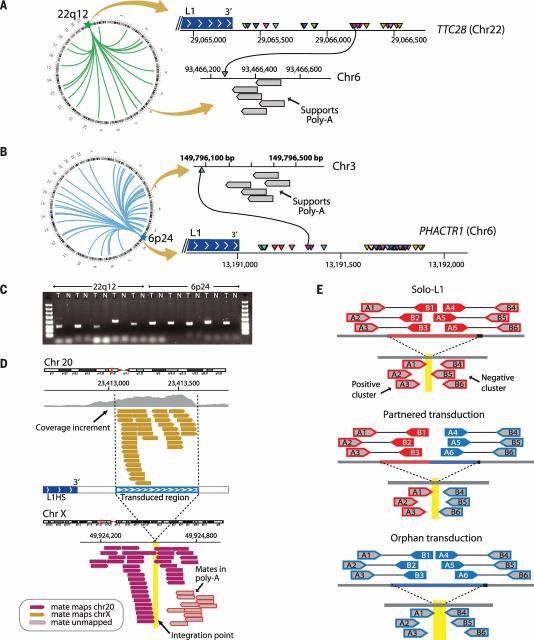
Fig. 1. Somatically acquired 3′ transductions are frequent in cancer genomes.
(A) Putative translocations involving 22q12 (TTC28) show characteristics suggestive of L1-mediated 3′ transduction. Breakpoints at 22q12 (triangles of different colors) are clustered immediately after a germline full-length element. On the other side of the breakpoint, there are reads whose pairs report the presence of poly(A) tails (gray boxes). (B) Breakpoints clustered in PHACTR1 just after a polymorphic L1 element not present in the reference genome. (C) PCR profiles showing the somatic acquisition of transductions from 22q12 and 6p24. T, tumor; N, normal. (D) The hallmarks of 3′ transduction. The donor-L1 locus at chromosome 20 shows coverage increment downstream of the element resulting from genome-wide amplification of the transduced material. Reads responsible for the coverage increment pair with different chromosomes (chromosome X illustrated). A cluster of reads around the breakpoint indicates the presence of a poly(A) tail. Other reads reveal the presence of target site duplication (not shown; details in table S3). (E) The strategy followed for somatic solo-L1 and transduction identification. The pipeline relies on the identification of two read clusters (i.e., positive and negative clusters) pointing to the same region of the genome where the somatic element is inserted.