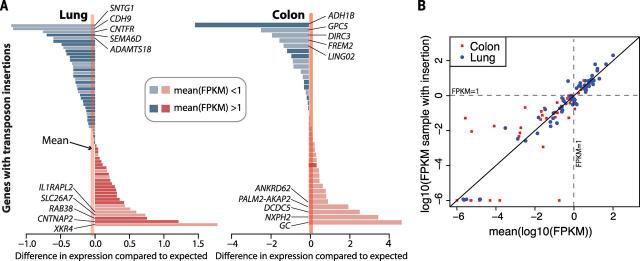
Fig. 7. Gene expression effects associated with L1 insertions.
(A) For lung and colon cancers, each bar represents the difference between the log10 (FPKM) for the target gene in the relevant sample compared to the average log10(FPKM) for other samples of that tumor type. FPKM, fragments per kilobase of transcript per million mapped reads. Genes with FPKM > 1 average expression have darker bars. (B) Scatter plot showing the data in (A).The y axis shows the log10(FPKM) for the target gene in the relevant sample, and the x axis shows the average log10(FPKM) for that gene for other samples of that tumor type. In expressed genes [mean log10(FPKM) > 0], the expression level in the affected sample is very close to the overall expression level of the gene in the corresponding tissue. Most large expression level differences occur at unex-pressed genes [mean log10(FPKM) < 0].