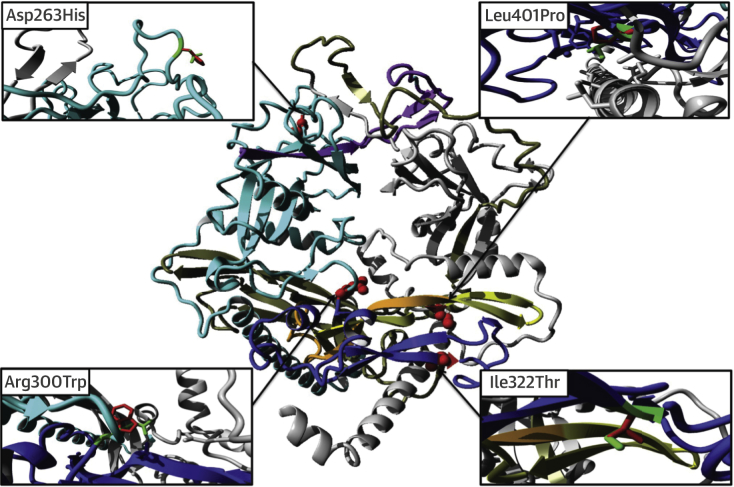
Figure 4.
Overview of the TGFB3 Dimer Model and Mutations
One monomer is shown in grey, with the other monomer in cyan (LAP domain) and blue (TGFB3 domain). Residues deleted by the p.Glu216_Lys251del mutation are in purple. Residues affected by the p.Leu386Argfs21* mutation are in yellow (note that this mutation also adds 21 different residues that cannot be modeled). Residues deleted by the p.Tyr365* mutation are shown in orange and yellow (note that this mutation deletes all residues following Tyr365). Residues deleted by mutation p.Asn235Metfs*11 are shown in grey-olive in the second monomer (note that this mutation also adds 11 different new residues, which cannot be modeled). The point mutations p.Asp263His, p.Arg300Trp, p.Ile322Thr, and p.Leu401Pro are shown in red with their side chains visible as red spheres. A detailed close-up of these mutations is shown in 4 extra panels. In these panels, the wild-type residue side chain is in green, whereas the mutant side chain is in red. For the p.Arg300Trp mutation, side chains of nearby negative residues are also shown. For the p.Leu401Pro mutation, nearby hydrophobic residues are shown.