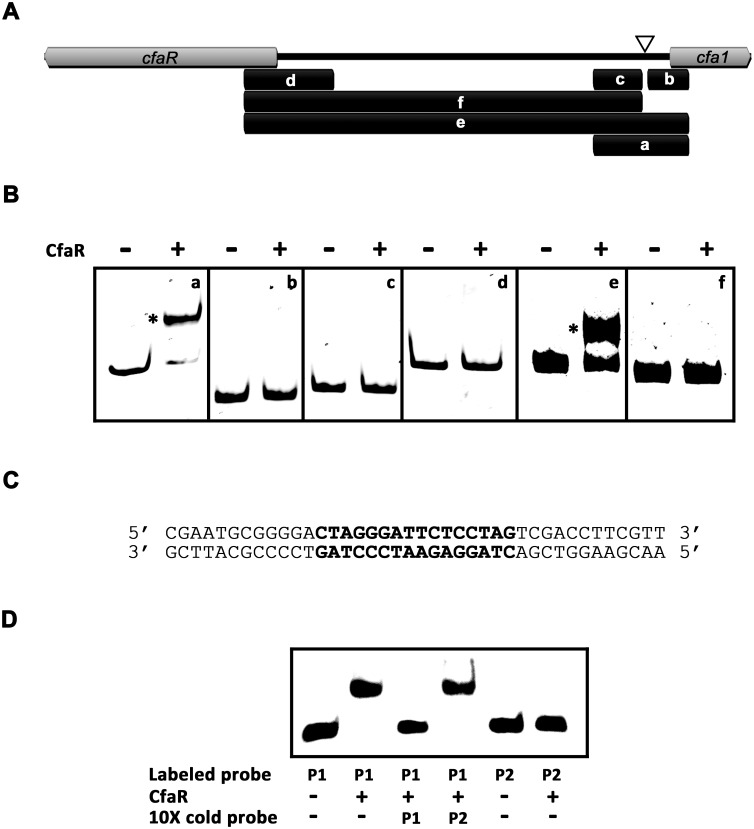
Fig 2. CfaRfull-HIS6 binds to a single site within the cfaR—cfa1 intergenic region.
(A) Map of the cfaR—cfa1 intergenic region showing the location of the DNA fragments (indicated by the black bars and labeled a—f) used for EMSAs. The position of the 16 bp palindrome identified upstream of cfa1 is indicated with the white triangle. (B) EMSA s for CfaRfull—HIS6 with the DNA fragments a—f. Reactions contained 50 ng of DNA with (result+) and without (-) CfaRfull—HIS6 protein (3.7 pmol). DNA-protein complexes observed are indicated with *. (C) Sequence of the 40 bp oligonucleotide P1 probe used for EMSAs. The 16 bp palindromic sequence identified upstream of cfa1 is shown in bold. (D) EMSA results for CfaRfull—HIS6 with the P1 oligonucleotide probe. Reactions contained 0.1 pmol of biotin-labeled probe with (+) and without (-) CfaRfull—HIS6 protein (2 pmol). Negative control reactions contained the 40 bp biotin-labeled oligonucleotide P2 probe in place of P1. In addition, competition assays were performed in which an excess (10×) of unlabelled (cold) probe (P1 or P2) was included in the reaction. DNA-protein complexes observed are indicated with *.