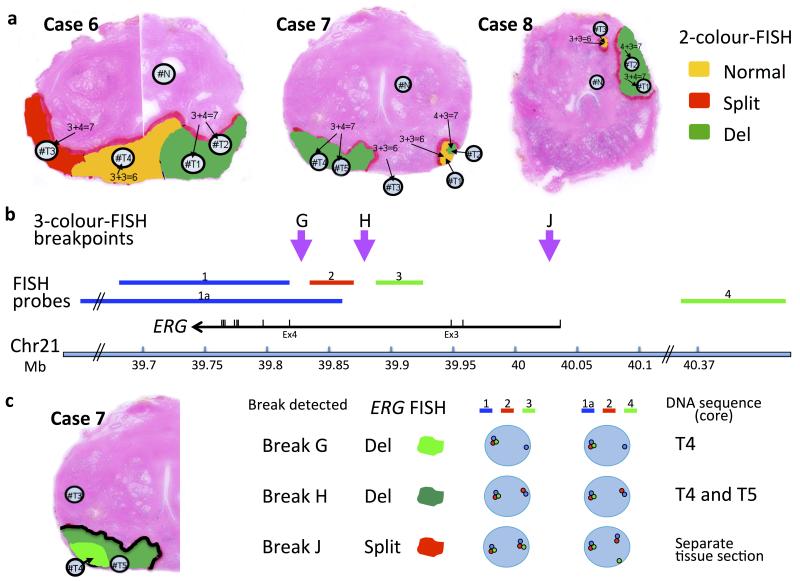
Figure 1.
Prostate samples chosen for whole-genome sequencing. a, ERG rearrangements determined by fluorescence in situ hybridization (FISH). Case 7 is a multifocal cancer containing two separate foci (T1/T2/T4/T5 and T3). Case 8 is also designated as a multifocal cancer,(nodules T1/T2, and T3). Yellow: un-rearranged normal ERG gene; Red, ERG gene split but both 3′ and 5′ ends retained; Green, ERG gene rearranged but only its 3′ end retained. Panels b and c: 3-colour FISH used to distinguish different ERG-locus translocation breakpoints in Case 7. b, Position of the three FISH probes: probe 1 (blue, BAC RP11-164E1, and probe 1a, BACs RP11-95G19, RP11-720N21, CTD-2511E13) was labeled in Aqua (Kreatech 415 Platinum Bright): probe 2 (red, fosmid G248P80319F5 37Kb) labeled with Cy3; and Probe 3 (green, fosmid G248P86592E2 38.5k, and probe 4, BACs RP11-372O17, RP11-115E14, RP11-729O4) labeled with FITC. The purple arrows represent the positions of ERG breakpoints detected in these experiments. For the precise position of the ERG breakpoints G and H see Table 2. c, Left: Tumor areas with ERG locus breaks G and H are indicated as light and dark green respectively. Break J was found in an adjacent prostate section not show in this figure. Right: representations of the ERG FISH patterns. Original FISH images are show in Supplementary Fig. 1. “Split” denotes that 5′ and 3′ ERG signals were separated but retained in the cell. “Del” indicates that 5′ ERG signals were lost from the cell, while 3′ ERG signals were retained.