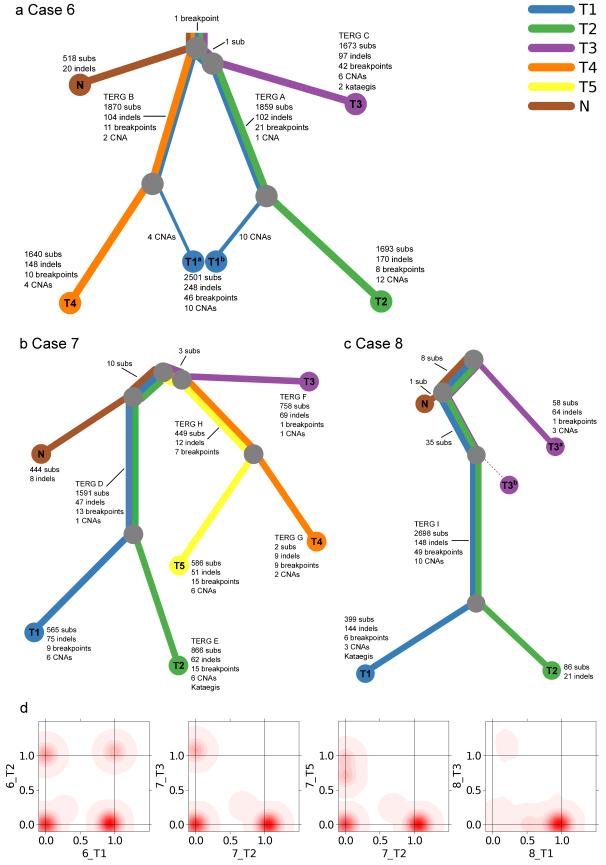
Figure 2.
Phylogenies of multi-focal prostate cancers. a-c, Phylogenies revealing the relationships between sample clones for each case. Each line is associated with a clone from a particular sample. The length of each line is proportional to the weighted quantity of variations on a logarithmic scale. The thickness of a line indicates the proportion the clone makes up of that sample i.e. 48%/52% for 6_T1 and 12%/88% for 8_T3. The minor clone of 8_T3b has no detected unique variants. 8_T3 contained 43 mutations present as a 12% subclone (T3a) shared with 8_T1/8_T2. In validation experiments 8_T3 did not contain any of the five ERG and TMPRSS2 rearrangements present in 8_T1/8_T2 (Table 2)) or mutations that were unique to 8_T1/8_T2 (10,000 depth) indicating that it represents an earlier clone of 8_T1/8_T2 seeded into tissue sample 8_T3. The various TMPRSS2-ERG translocations are indicated by their TERG ID (Table 2). d, Example 2D density plots showing the posterior distribution of the fraction of cells bearing a mutation in two samples. The fraction of cells is modeled using a Bayesian Dirichlet processes. These plots illustrate samples that have shared clonal mutations (6_T1/6_T2), and branched (unrelated) mutations (7_T2/T_T3). There are two examples of samples with a subclone. 7_T2/7_T5 has a peak at (0,0.72), which represents subclonal mutations in 72% of cells in 7_T5 that have occurred only in this sample, after divergence from the other samples. Similarly, 8_T1/8_T3 has a peak at (0.54,0), representing subclonal mutations in 54% of cells in T1 only.