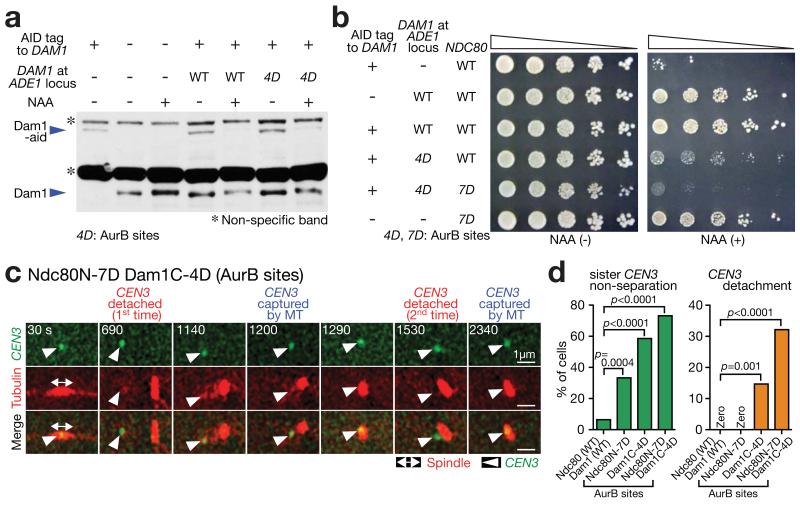
Figure 5. Aurora B phospho-mimicking Dam1 and Ndc80 mutants show repeated turnover of KT–MT attachments.
(a) Western blotting (with Dam1 antibody) showing Dam1-aid depletion and Dam1 protein (wild-type or Dam1C-4D[AurB]) expression from an additional DAM1 construct. Cells were incubated with or without auxin NAA for 4 h in methionine dropout medium. In Dam1C-4D[AurB], four Aurora B target sites (S257, S265, S292 and S327 18) were replaced with aspartates at the Dam1 C-terminus. A full scan of the western blot is shown in Supplementary Fig 6.
(b) Serially diluted cells were incubated with or without auxin NAA. In Ndc80 N-7D[AurB], seven Aurora B target sites (T21, S37, T54, T71, T74, S95 and S100 19) were replaced with aspartates at the Ndc80 N-tail.
(c, d) NDC80+ DAM1+ (T9530), ndc80N-7D[AurB] (T11102), dam1C-4D[AurB] dam1-aid (T11326) and ndc80N-7D[AurB] dam1C-4D[AurB] dam1-aid (T11452) cells with TIR PGAL-CEN3-tetOs TetR-3×CFP GFP-TUB1 PMET3-CDC20 were treated with α factor for 3 h and released to fresh media with methionine (for Cdc20 depletion) in the presence of NAA (to deplete Dam1-aid) and glucose (CEN3 under the GAL promoter was always active). From 90 min after release, images were acquired every 30 sec. c shows a representative cell with ndc80N-7D[AurB] dam1C-4D[AurB] (0 sec: start of image acquisition). d displays percentage of cells showing sister CEN3 non-separation (left) and CEN3 detachment from the spindle (usually followed by reattachment; right); n= 62, 48, 75 and 56 cells were analyzed (from left to right). Experiments were performed twice (statistics source data are shown in Supplementary Table 2) and a representative experiment is shown here. p-values (two tailed) were obtained by Fisher’s exact test.