Figure 1. Biochemical basis of ΔFosB’s unique stability.
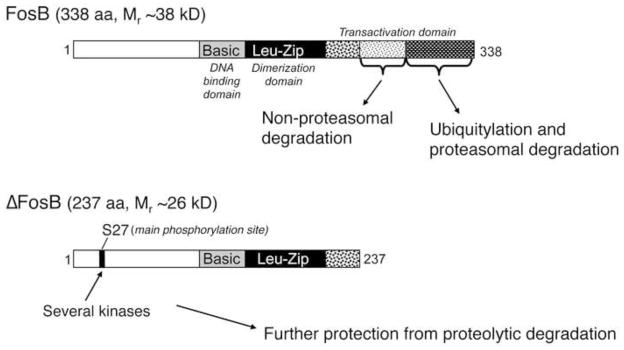
ΔFosB and FosB are encoded by the FosB gene. ΔFosB is generated by alternative splicing and lacks the C-terminal 101 amino acids present in FosB. Two mechanisms are known that account for ΔFosB’s stability. First, ΔFosB lacks two degron domains present in the C-terminus of full length FosB (and found in all other Fos family proteins as well). One of these degron domains targets FosB for ubiquitylation and degradation in the proteasome. The other degron domain targets FosB degradation by a ubiquitin- and proteasome-independent mechanism. Second, ΔFosB is phosphorylated by several protein kinases at its N-terminus which further stabilizes the protein. From Nestler, 2008 with permission.
