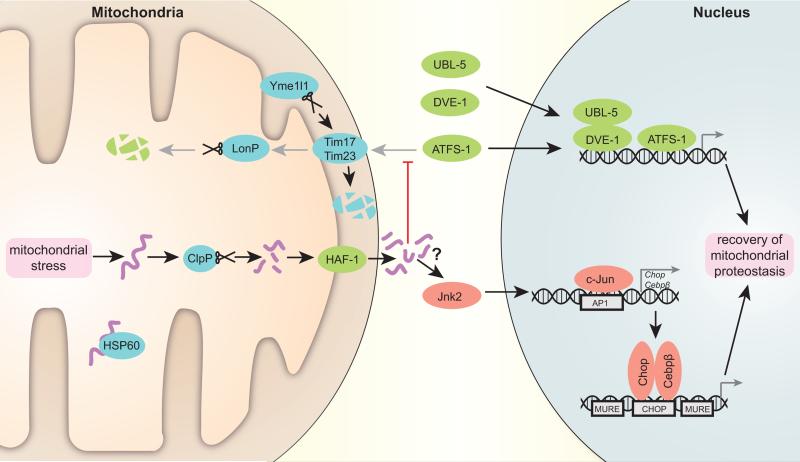
Figure 1. Scheme depicting the transcriptional regulation of the UPRmt.
Accumulating unfolded proteins, unassisted by chaperone Hsp60 in stressed mitochondria, are digested by the protease Clpp. The resulting peptides are transported through the double mitochondrial membrane into the cytosol. These peptides presumably stop mitochondrial import, which is also negatively affected by specific degradation of Tim17 component of the translocation pore by protease Yme1l1. As a result, C. elegans transcription factor ATFS-1, which in normal conditions is translocated to mitochondria and degraded by protease LonP, moves into the nucleus together with UBL-5 and DVE-1 to activate a reparative transcriptional program. In mammals, Jnk2 triggers c-Jun binding to AP1 sites, leading to the activation of Chop and Cebpβ transcription. Subsequently, Chop and Cebpβ dimers bind to CHOP sites flanked by MUREs and induce UPRmt target gene transcription. Proteins characterized in C. elegans are marked in green, fly and mammalian system proteins in red and proteins conserved in all the systems are noted in blue (mouse nomenclature is used).