Figure 4.
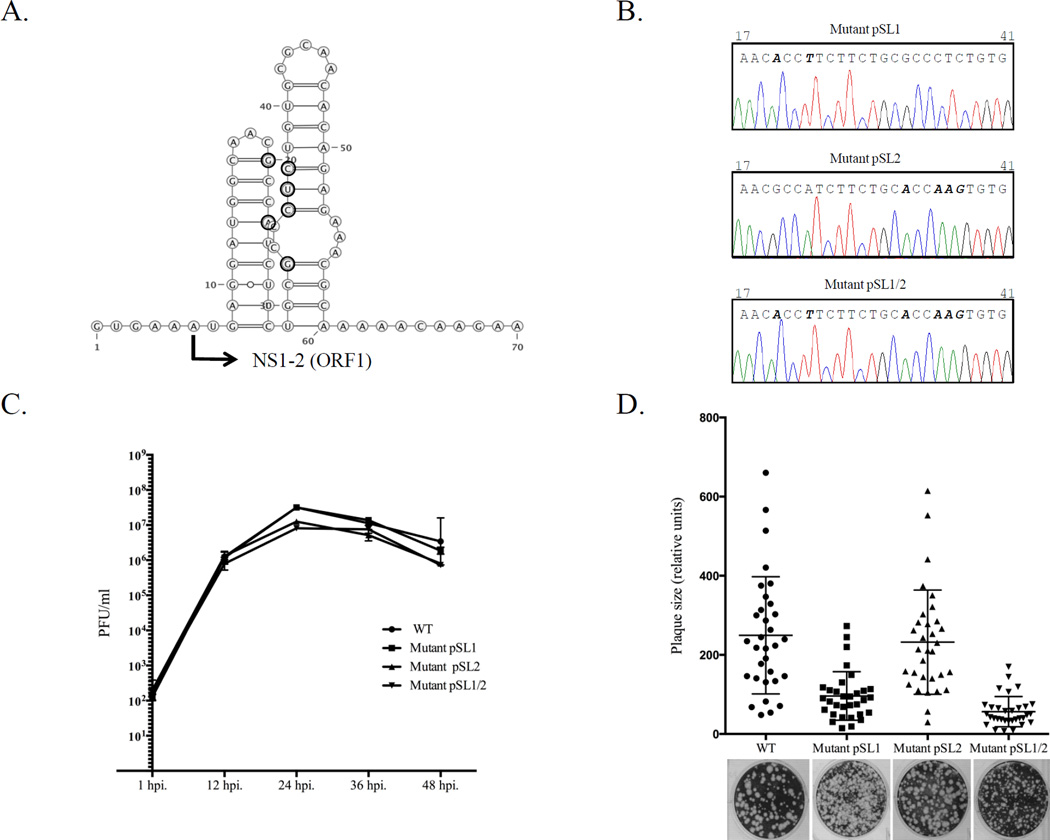
Generation of genetically-modified MNV. (A) Site-directed mutagenesis was employed to introduce silent mutations in the beginning of the NS1-2 gene. The corresponding positions are indicated with bold circles and the beginning of the NS1-2 gene (ORF1) is marked with arrow. The introduced changes could alter the RNA secondary structure predicted for this part of the MNV genome (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). (B) Comparison of the genome sequence (nts 17-41) determined for the recovered SL1, SL2, and SL1/2 mutant viruses at passage 3. Mutations at positions 20 and 23 (SL1), 32 and 35–37 (SL2) and 20, 23, 32, and 35–37 (SL1/2) are shown in bold. (C) Growth properties of mutant viruses were compared with those of the wild-type (wt) virus. Confluent monolayers of RAW264.7 cells were infected with wt or mutant viruses at a multiplicity of infection of 0.05. Cell culture fluid was harvested 1, 12, 24, 36, and 48 h post infection. Virus titer was determined by end point titration in a plaque assay. (D) Recovered mutant viruses were assayed for their ability to form plaques in RAW264.7 cells. The relative size of 32 randomly selected plaques was plotted.