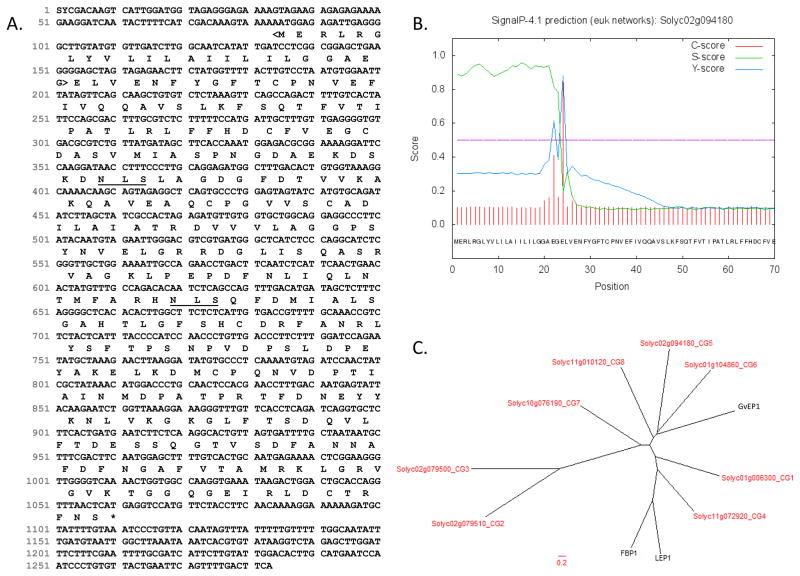
Figure 2. Sequence analyses of the tomato CG5.
(A) Full-length cDNA and predicted protein sequence are from iTAG2.3 (accessed 3/5/14). N-glycosylation sites (underlined) were predicted by NetNGlyc1.0. The N-terminal signal sequence (<bracketed>) and (B) cleavage site were predicted by SignalP 4.0 (Petersen et al. 2011). (C) Phylogenetic analysis of EP proteins and tomato CGs was performed. Predicted protein sequences of tomato CGs were aligned with EP proteins from grapevine (GvEP1; Jackson et al. 2001), lupin (LEP1; Price et al 2004), and French bean (FBP1; Wojtaszek et al. 1997). Multiple sequence alignment was performed by ClustalW using a GONNET weight matrix with a gap penalty of 10 and an extension penalty of 0.05. TreeDyn was used to make the radial dendrogram. Tomato CGs are shown in red.