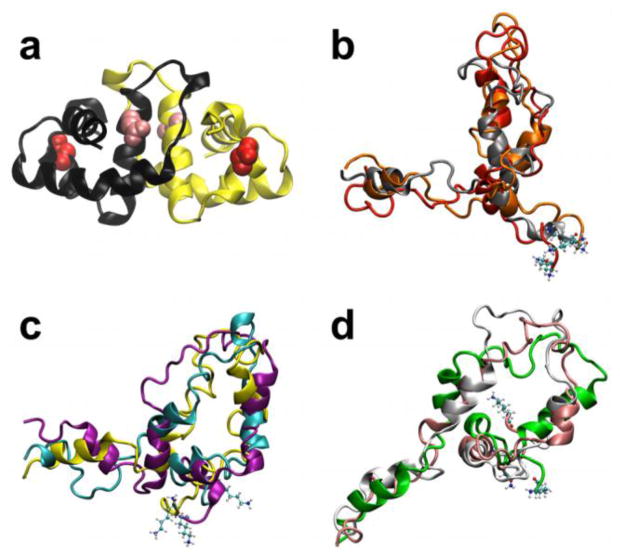
Figure 1.
Folded dimer and unfolded monomer structures of HdeA. (a) Crystal structure of the HdeA homodimer (PDB ID: 1BG8)47. The acidic residues that exhibit the most significant destabilizing (Asp20, red) and stabilizing (Glu37, pink) effects on HdeA upon protonation at low pH are denoted as van der Waals spheres. (b–d) Representative conformers of the unfolded ensemble of the HdeA monomer from clustering of the coarse-grained ensemble. The corresponding clusters have the following fractional population sizes: (b) 0.24 (c) 0.41, and (d) 0.35. The N-terminal residue in each unfolded conformer is denoted in CPK representation.