Figure 5.
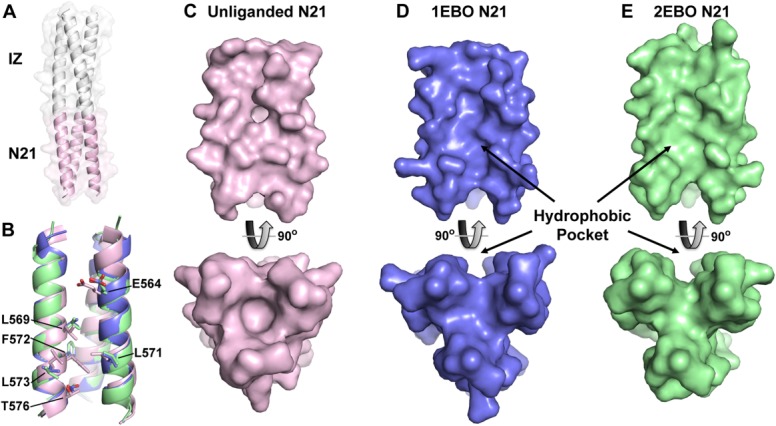
Crystal structure of eboIZN21. (A) Cartoon rendering with a semitransparent surface of the unliganded eboIZN21 structure. The IZ trimerization domain (white) and N21 region (pink) are indicated. The N21 region of available ebolavirus N-trimer structures is shown in isolation in panels B–E. (B) Overlay of the N21 region of the unliganded structure with the N21 region of the two previously solved ebolavirus GP2 core structures containing C-peptide (PDB IDs: 1EBO and 2EBO shown as blue and green, respectively. This color scheme is maintained in panels C–E). Residues that line the N21 groove and have significantly different rotamer conformations in the unliganded structure are shown as sticks and labeled. These residues occupy some of the equivalent space occupied by C-peptide (not shown) in the liganded structures resulting in a less prominent hydrophobic pocket when viewed (in subsequent panels) as a surface. (C) Surface representation of the unliganded N21 region. The bottom panel is the view of N21 from the bottom along its threefold axis and is rotated approximately 90° as compared to the top panel. (D, E) Similar views to (C) of the N21 region from the structures containing C-peptide. The prominent hydrophobic pocket in the 1EBO and 2EBO structures appears to be induced by ligand binding since the pocket is nearly absent in the unliganded structure.
