Figure 1.
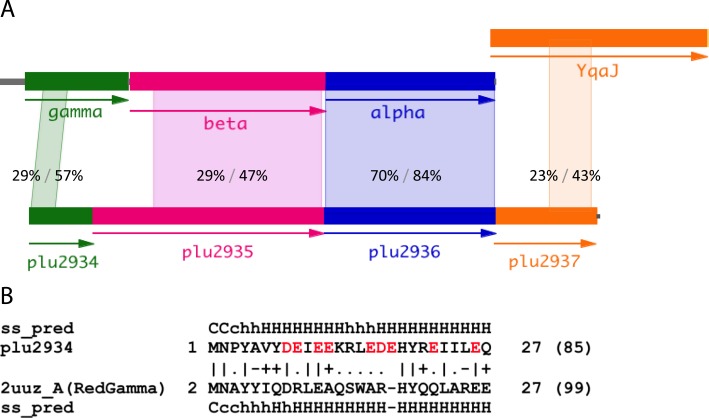
Bioinformatics analysis of Plu2934-6. (A) Operon architecture of λ Red and Plu2934-Plu2937. Photorhabdus prophage Plu2934, Plu2935 and Plu 2936 are related to Redγ (gamma), Redβ (beta) and Redα (alpha), respectively, encoded by λ phage. Plu2937 shares similarity with YqaJ encoded by the Bacillus subtilis prophage. The amino acid sequences were compared with NCBI protein Blast analysis. The extent, position, percentage identity and similarity are displayed between the homology regions. (B) Plu2934 protein homology detection of known structures using the HHpred interactive server. The Redγ structure is taken from PDB entry 2uuz chain A and exhibits alpha-helical structure. The negatively charged residues aspartic acid (D) and glutamic acid (E) are marked in red. ss_pred: secondary structure prediction by PSIPRED; H/h: alpha-helix; C/c: coli; upper case: high probability; lower case: low probability; quality of column match (bad: −; neutral:.; good +; very good |).
