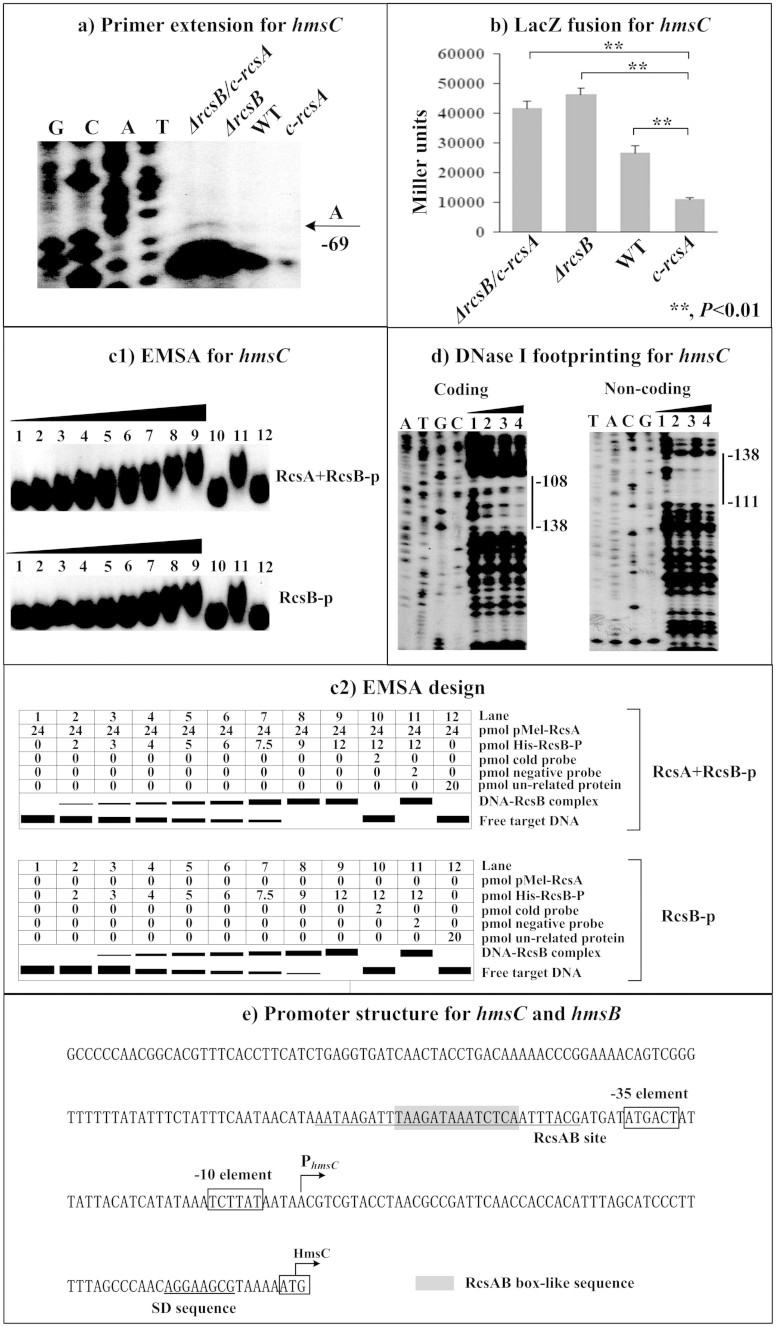
Figure 2. RcsAB-dependent expression of hmsCDE.
(a) Primer extension. The relative mRNA levels of hmsC in indicated strains were determined by primer extension. The Sanger sequence ladders (lanes G, C, A, and T) and the primer extension products of hmsC were analyzed with an 8 M urea-6% acrylamide sequencing gel. The transcription start site of hmsC was indicated by arrow with nucleotide A, and the minus number under arrow indicated the nucleotide position upstream of hmsC start codon. (b) LacZ fusion. The hmsC:lacZ transcriptional fusion vector was transformed into in indicated strains, and then hmsC promoter activities (miller units of β-galactosidase activity) were determined in bacterial cellular extracts. (c) EMSA. The radioactively labeled DNA fragments were incubated with indicated purified proteins and then subjected to a native 4% polyacrylamide gel electrophoresis. (d) DNase I footprinting. Labeled coding or non-coding DNA probes were incubated with indicated purified proteins and then subjected to DNase I digestion. The digested DNA samples were analyzed in an 8 M urea-6% polyacrylamide gel. The footprint regions were indicated with vertical bars. Lanes C, T, A, and G represented Sanger sequencing reactions. The DNA-binding of His-RcsB-p in presence of MBP-RcsA (involved in EMSA and DNase I footprinting) and that of His-RcsB-p alone (in EMSA) were tested. (d) Promoter structure. Shown were with translation/transcription starts, core promoter −10 and −35 elements, SD sequences, RcsAB sites, and RcsAB box-like sequences.