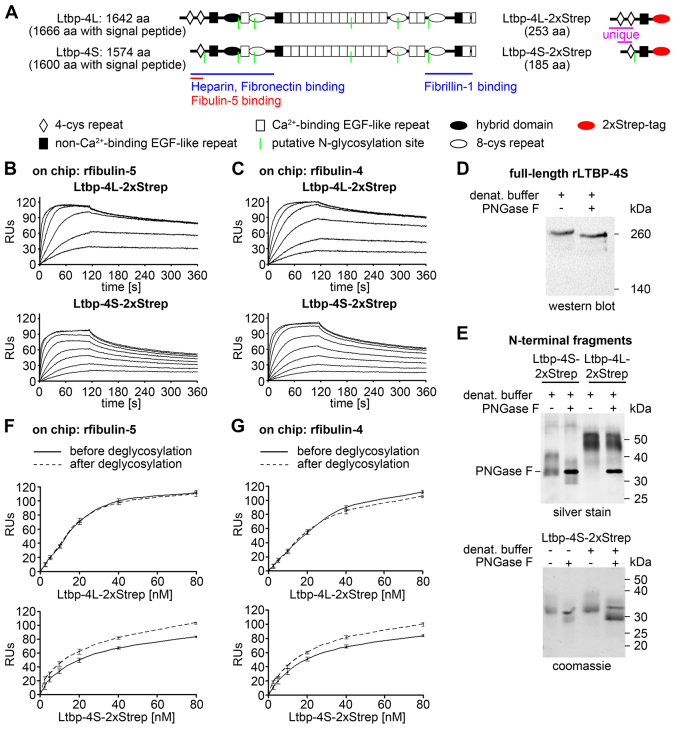
Fig. 4.
Interaction studies of the Ltbp-4L and Ltbp-4S N-terminal regions with full-length fibulin-4 and fibulin-5. (A) Domain structure of full-length Ltbp-4L and Ltbp-4S and the recombinantly expressed Ltbp-4L (Ltbp-4L-2xStrep) and Ltbp-4S (Ltbp-4S-2xStrep) N-terminal fragments. The full-length proteins consist of 4-cystein (4-cys) repeats (white rhombi), non-Ca2+-binding EGF-like repeats (black rectangles), Ca2+-binding EGF-like repeats (white rectangles), hybrid domains (black ellipses) and 8-cystein (8-cys) repeats (white ellipses). The N-terminal fragments consist of two (Ltbp-4L-2xStrep) or one (Ltbp-4S-2xStrep) unique 4-cys repeats, the common non-Ca2+-binding EGF-like repeat and a C-terminal 2xStrep tag (red ellipses). Binding sites for ECM proteins, putative N-glycosylation sites (green lines) as well as the amino acid (aa) lengths are indicated. (B,C) Sensorgrams from surface-plasmon resonance interaction experiments showed a stronger binding affinity of Ltbp-4L-2xStrep (0–320 nM) ‘flown’ over immobilized recombinant full-length fibulin-5 (B; rfibulin-5) or immobilized recombinant full-length fibulin-4 (C; rfibulin-4) compared to Ltbp-4S-2xStrep (0–80 nM) flown over immobilized rfibulin-5 (B) or rfibulin-4 (C). The results are expressed as resonance units (RUs; n=2). (D) Deglycosylation digest with PNGase F of denatured recombinant full-length human LTBP-4S (rLTBP-4S) showing that there is a shift towards lower molecular mass positions. (E) Upper panel: deglycosylation of Ltbp-4L-2xStrep and Ltbp-4S-2xStrep. Ltbp-4L-2xStrep was unaffected, whereas Ltbp-4S-2xStrep showed a shift towards lower molecular weight positions. Lower panel: Ltbp-4S-2xStrep was digested with PNGase F under native (left lanes) and denaturing (right lanes) conditions. Both conditions resulted in a shift towards lower molecular weight positions. (F,G) After digest under non-denaturing conditions Ltbp-4L-2xStrep and Ltbp-4S-2xStrep were both able to bind to rfibulin-4 and -5 immobilized on a Biacore chip. Ltbp-4L-2xStrep binding was not affected, whereas Ltbp-4S-2xStrep showed an increase in binding of 15% to 20% after deglycosylation. The continuous lines represented the response before and the dashed lines after deglycosylation (n=2).