Figure 6.
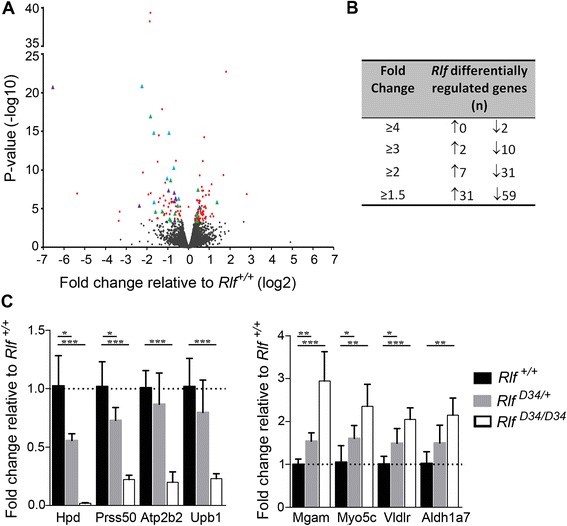
Loss of Rlf influences transcription in the fetal liver and is associated with alterations in epigenetic state. (A) Volcano plot depicting the results of RNA-seq analysis of RNA from E14.5 livers of Rlf +/+and Rlf MommeD28/MommeD28 mice. Three independent biological replicates were analysed per genotype. Significantly differentially expressed genes are presented as red, blue, green or purple data points, p < 0.05. Genes that have either a Rlf-DMR or a Rlf-dependent change in H3K4me1 occupancy within 50 kb of their TSS are highlighted by green and purple triangles, respectively. Blue triangles highlight genes with a change in both DNA methylation and H3K4me1 occupancy. Black data points represent genes whose expression was not significantly altered by loss of Rlf. ( B ) Table showing the number of genes significantly differentially expressed in Rlf MommD28/MommeD28 E14.5 livers relative to Rlf +/+ controls at various fold-change cut-offs (p < 0.05). ↑ and ↓ indicates increased or reduced expression, respectively, in Rlf MommeD28/MommeD28 homozygotes. (C) Quantitative real-time RT-PCR analysis of putative Rlf-regulated genes identified via RNA-seq. Analyses used RNA extracted from E14.5 fetal livers of wild-type mice and an independent null Rlf mutant mouse line, MommeD34, see Introduction. Mean ± SEM is present for at least six individuals per genotype. Statistical significance was determined via a t-test *p < 0.05, **p < 0.005.
