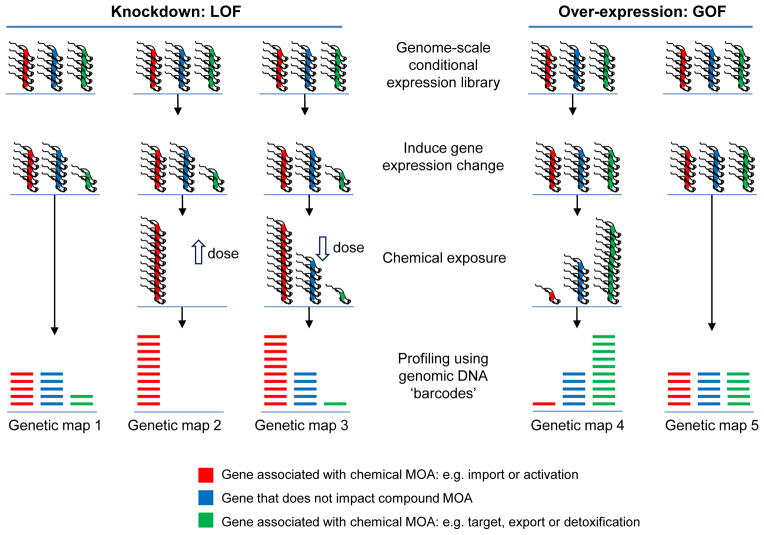
Figure 3.
Chemical genetic screens. Genome profiling using knockdown or over-expression of every reading frame to probe mechanism of action (MOA) and interaction, most of what is shown is yet to be implemented for parasites. Open arrows indicate high and low-dose chemical exposure. The schematic illustrates highly parallel co-culture fitness tests following conditional gene expression changes, either loss-of-function (LOF) or gain-of-function (GOF), and chemical exposure. Complex populations with protein expression down or upregulated are grown competitively and then quantitatively assessed using a read-out, typically generated using DNA sequencing and mapping to a reference genome. In the case of green genes, loss of fitness in map 1 means that it is more likely that this gene is a target. A more moderate knockdown could be used to increase the differential read-out, due to drug-RNAi synergy, in maps 1 and 3. Genes indicated in red and blue may also be associated with a fitness cost when knocked down (or occasionally when over-expressed, neither indicated). In these cases, differential representation may still reveal the chemical interaction indicated for red genes in maps 1 to 3. Conversely, genes indicated in green may not be associated with a fitness cost when knocked down if they are not targets; these could be dispensable pumps involved in drug export for example. Trypanosomes are shown but the principles are similar for other cell types. ‘Functional variomics’ screens, combining both mutations and over-expression, have recently been applied to budding yeast (156).