Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that play important post-transcriptional regulatory roles in a wide range of biological processes. They are fundamental to the normal development of cells, and evidence suggests that the deregulation of specific miRNAs is involved in malignant transformation due to their function as oncogenes or tumor suppressors. We know that miRNAs are involved in the development of normal B-cells and that different B-cell subsets express specific miRNA profiles according to their degree of differentiation. B-cell-derived malignancies contain transcription signatures reminiscent of their cell of origin. Therefore, we believe that normal and malignant B-cells share features of regulatory networks controlling differentiation and the ability to respond to treatment. The involvement of miRNAs in these processes makes them good biomarker candidates. B-cell malignancies are highly prevalent, and the poor overall survival of patients with these malignancies demands an improvement in stratification according to prognosis and therapy response, wherein we believe miRNAs may be of great importance. We have critically reviewed the literature, and here we sum up the findings of miRNA studies in hematological cancers, from the development and progression of the disease to the response to treatment, with a particular emphasis on B-cell malignancies.
Keywords: MicroRNA, B-cell, Differentiation, B-cell malignancies, Drug response
INTRODUCTION
MicroRNAs (miRNAs) are central players in the regulation of cellular processes from proliferation and differentiation to metabolism and apoptosis [1]. They are fundamental to the normal development of cells and evidence implies that miRNA deregulation is involved in the pathogenesis of human diseases. Translational research has identified candidate miRNA biomarkers for use in diagnosis and treatment guidance in a broad range of diseases, including hematological cancers [2, 3].
Most non-Hodgkin lymphomas (NHL) are considered to be derived from germinal center (GC) cells based on histological features, immunohistochemical characteristics, and immunoglobulin (Ig) gene rearrangements. Implementation of global gene expression profiling (GEP) has allowed for a more comprehensive phenotype-based assignment to a cell of origin (COO) classification, reminiscent of the normal B-cell differentiation program. In diffuse large B-cell lymphoma (DLBCL), GEP-based COO classification recognizes three main subclasses: GC B-cell-like (GCB) DLBCL, which represents transformed GC centroblasts (CB) [4, 5]; activated B-cell-like (ABC) DLBCL, which resembles post-GC plasmablasts [6]; and primary mediastinal large B-cell lymphoma (PMBCL), thought to originate from a thymic post-GC B-cell [7-9]. In agreement with different messenger RNA (mRNA) profiles, distinct miRNA signatures are associated with molecular subclasses of DLBCL [10-13]. In support of the GEP-based subclassifications, there are observations of recurrent genetic abnormalities with subclass-specific distributions and activation of distinct signaling pathways [14]. Thus, each naturally occurring differentiation subset of B-cells has intrinsic gene and miRNA phenotypic profiles granting them specific abilities to further differentiate, respond to stimuli, and survive when exposed to drugs. These features can be partly maintained when a malignant condition initiates in a cellular differentiation-specific compartment. The malignancy then either develops by expansion of that same end-stage compartment or differentiates into a more mature one. We believe the classification of NHL can be further refined using GEP and miRNA expression profiles determined in normal B-cell differentiation subsets isolated from healthy human secondary lymphoid tissue [15, 16]. This idea has prompted us to look into the translational research of miRNAs and their use in the characterization of normal and malignant B-cell subsets.
This review will present examples of the most recent miRNA translational studies. We will focus on the impact of miRNAs on the development and progression of hematological malignancies, especially from the B-cell lineage, and on their potential as biomarkers of prognosis and response to chemotherapy.
BIOGENESIS AND ACTIVITY OF miRNAs
miRNA biogenesis
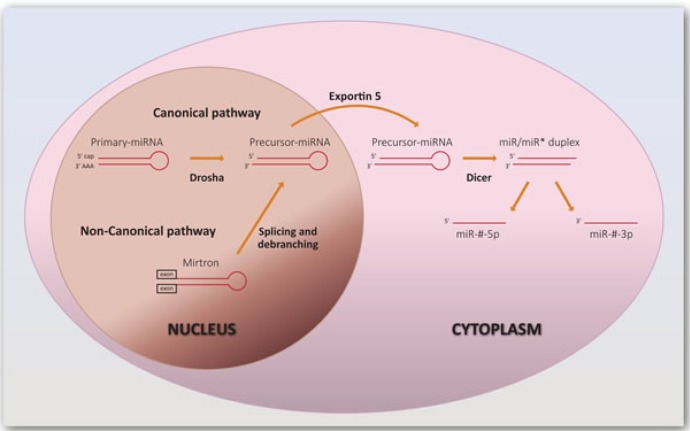
The synthesis of mature and fully competent miRNAs is designated miRNA biogenesis, a process that begins in the nucleus and is completed in the cytoplasm (Figure 1). In the canonical biogenesis pathway, intergenic miRNA genes are transcribed by RNA polymerase II into 5′-capped and 3′-polyadenylated molecules, the primary-miRNAs [17]. These are processed by a protein complex comprising Drosha and DiGeorge syndrome critical region gene 8 (DGCR8), generating intermediates of approximately 70 nucleotides (nts) called precursor-miRNAs, which contain an approximately 2 nts long 3′ overhang [18]. Precursor-miRNAs are transported to the cytoplasm by Exportin 5 [19] and cleaved by a complex that includes the RNAse enzyme Dicer and TAR RNA-binding protein (TRBP), resulting in a miRNA duplex [20-23]. The duplex is separated originating mature miRNAs, which are 20 to 25 nts long single-stranded RNAs. One of the strands, the passenger strand, is usually degraded and the other, called the guide strand, is loaded into the RNA-induced silencing complex (RISC) with Argonaute (Ago) proteins, enabling the targeting of mRNAs through complementary base pairing [24].
Figure 1. Biogenesis of miRNAs.
The synthesis of miRNAs via the canonical pathway starts with transcription of miRNA genes by RNA polymerase II, which results in primary-miRNAs. These are processed by Drosha and DiGeorge syndrome critical region gene 8 (DGCR8), generating precursor-miRNAs which are transported to the cytoplasm by Exportin 5 and cleaved by Dicer and TAR RNA-binding protein (TRBP). The resulting duplex is separated, generating mature miRNAs. MiRNAs can also arise from splicing through the non-canonical pathway. The designation of miRNAs includes the term “miR” preceding a number attributed sequentially. Similar sequences can have the same number but a different suffix (number or letter). Letters defining the species are added as prefixes, such as hsa for Homo sapiens. Additionally, they can be designated miR-#-3p or miR-#-5p depending on which arm of the precursor structure the leading strand is located [168].
Another less common process of synthesizing miRNAs arises from splicing. This non-canonical pathway leads to the formation of mirtrons, which are miRNAs encoded in introns with hairpin-forming potential. They are not cleaved by Drosha, but are spliced out directly by the spliceosome complex, and they enter the biogenesis at the precursor stage [25, 26].
miRNA activity
A single miRNA can target hundreds of mRNAs [27], and every mRNA can be regulated by several miRNAs [28]. This confers onto these small RNAs a broad scope of action, affecting a multitude of cellular transcripts and pathways.
The miRNA guide strand can be either the 3′ or the 5′ of the precursor stem-loop structure, meaning that both sequences have the potential to become mature miRNAs. Strand selection for loading into the RISC can be either protein-mediated or based on thermodynamic stability [29]. Both strands are capable of functional activity, and the passenger strand, also called miRNA* or complementary strand, has also been documented to be involved in human disease [30-33].
Mature miRNAs bind to their target mRNA through partially or fully complementary base pairing primarily in the 3′ untranslated region (UTR) of the mRNA [34]. Most frequently, this induces inhibition of translation or mRNA degradation, which can be mediated by P-bodies [35]. However, miRNAs can also bind to their targets and act in other ways, as exemplified by miR-223, which binds to the promoter region of NFI-A and inhibits its transcription, reducing both mRNA and protein levels [36], and by the de novo DNA methylation of the promoter region of hoxd4 by miR-10a, resulting in transcriptional downregulation [37]. Regulatory functions through the targeting of the open reading frame of mRNAs mediating repression have also been reported [38-41].
MiRNAs can also activate translation and help stabilize viral mRNA, such as in the function of miR-122 towards the hepatitis C virus [42-45]. Additionally, they can be directly regulated by other RNAs, as suggested by the competing endogenous RNA (ceRNA) hypothesis from Paolo Pandolfi's group [46]. In summary, the better-known mechanism of action of miRNAs is the degradation or inhibition of translation of their target mRNA(s). However, miRNAs can also upregulate mRNA translation, can be modulated by mRNAs and other non-coding RNAs, and can be responsible not only for post-transcriptional but also transcriptional regulation.
B-CELL DIFFERENTIATION
B-cells undergo a stepwise differentiation process initiating from hematopoetic stem cells located in the bone marrow, where they differentiate into precursor B-cells [47]. This maturation process is characterized by a rearrangement of the V (variable), D (diversity), and J (joining) gene segments of the Ig genes. When the B-cell antigen receptor (BCR), comprising two identical heavy-chain and two light-chain Ig polypeptides, has been tested for auto-reactivity, the naïve B-cells leave the bone marrow and migrate via the blood to the secondary lymphoid tissues. Here, GCs are formed upon an encounter between the BCR and a foreign antigen [48-50]. In the GC a dark and a light zone can be distinguished. The dark zone consists mainly of proliferating CB undergoing somatic hypermutation whereas centrocytes (CC) are located in the light zone. The differentiation of CB and CC includes several rounds of migration between the dark and the light zones. A re-encounter between the B-cell and the antigen in a T-cell and follicular dendritic cell-dependent manner within the light zone ensures increased affinity between the Ig and the antigen. Following optimal antibody selection, a shift in the effector function by class switch DNA recombination (CSR) takes place in the CC in the light zone. The B-cells then leave the GC as memory B-cells or plasmablasts [49, 51, 52].
MiRNAs in B-cell differentiation
MiRNAs are fundamental to the development of blood cells, capable of regulating almost every stage of hematopoiesis [53] with lineage and differentiation-specific expression [54]. They are important determinants of B-cell maturation [55], and different stages of normal B-cell differentiation are characterized by different miRNA expression profiles [56-58].
When the expression of Dicer or members of the Ago family are removed, the synthesis of mature miRNAs in mouse models is impaired and B-cell differentiation is affected, highlighting the importance of miRNAs in the formation of B-cells [59]. When Dicer is ablated, early transition from pro-B to pre-B-cells [55], formation of GC B-cells [60], and terminal B-cell differentiation [61] are blocked. Thus, it is clear that antigen-dependent activation is not the sole driver of the formation of effector B-cells; their maturation is also highly dependent on the regulatory role of miRNAs.
Selectively targeting and manipulating the expression of miRNAs allowed the determination of their function at specific steps of B-cell differentiation. One of the first miRNAs identified in this manner was miR-181 (present name miR-181a-5p). Ectopic overexpression of this miRNA in hematopoietic stem-progenitor cells caused an increased fraction of B-cells in both tissue culture differentiation assays and adult mice [62]. The fact that miR-181a-5p is highly expressed in early human CD34+ hematopoietic stem-progenitor cells [63] and is downregulated in pre-BII [57] is indicative of an important function in early B-cell development. Additionally, it is predicted to inhibit differentiation of all hematopoietic lineages in an integrative bioinformatics analysis of miRNA and mRNA expression in human stem-progenitor cells from bone marrow and peripheral blood [63]. These findings are in accordance with studies in human immature precursor B-cell subsets, where miR-181a-5p was found to inversely correlate with the differentiation inhibitor ID2 mRNA, supporting a regulatory role in early differentiation of B-cells [57].
Like miR-181a-5p, manipulation of miR-150 in ectopic expression studies has provided insight into its role in normal B-cell differentiation. When miR-150 is overexpressed in murine hematopoietic stem-progenitor cells, the transition from pro-B to pre-B-cell is blocked and mature B-cells are reduced in numbers. MiR-150-knockout mice displayed a fourfold expansion of mature B1 cells, which are innate immune cells, combined with the enhanced apoptosis of pro-B-cells [64, 65]. Therefore, in normal B-cell differentiation, expression of miR-150 is high in the mature B-cells and relatively low in the immature ones, and is especially decreased at the pro-B to pre-B-cell transition [53, 64, 66]. The primary target of miR-150 is c-Myb, a transcription factor controlling multiple steps of B-cell development. Manipulation of the expression of miR-150 fits well with affected c-Myb function because the transition from pro-B to pre-B-cells requires c-Myb, which is consequently indirectly necessary for the generation of mature B-cells [65].
MiR-155 is highly expressed in GC B-cells and relatively low levels are observed in hematopoetic stem cells and mature B-cells [63, 67, 68]. It plays an important role in the control of the GC reaction, as documented by an increased number of GC B-cells in transgenic mice overexpressing this miRNA compared to knockout mice [69]. MiR-155-knockout mice had a reduced memory B-cell response when challenged by antigen immunization [69, 70] and B-cells lacking miR-155 presented reduced levels of high-affinity IgG1 antibodies [71]. Involvement of miR-155 in the regulation of activation-induced cytidine deaminase expression during affinity maturation and CSR in the GC, together with miR-181b, further support the role of miR-155 in B-cell differentiation [72, 73].
The cluster miR-17-92 has also been shown to be expressed in GC cells, and miR-125b, miR-9, and members of the miR-30 family are overexpressed in the GC compared to plasma cells, showing evidence of their involvement in the differentiation of B-cells [11, 58].
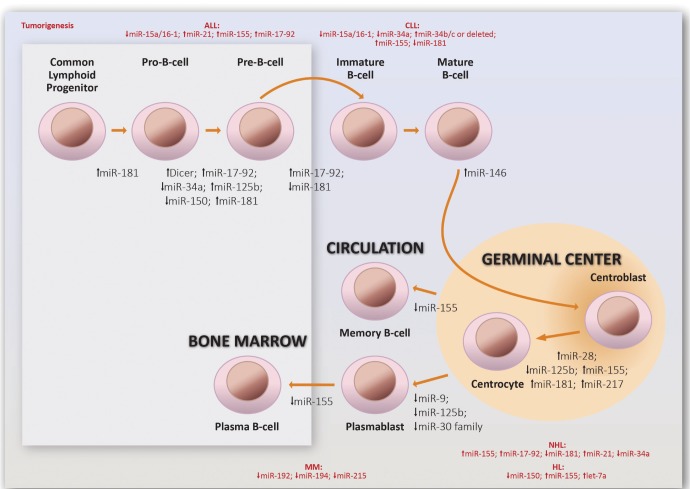
These are clear examples of how distinct B-cell subsets express different patterns of miRNAs, which we believe contributes greatly to their ability to differentiate. Figure 2 illustrates examples of miRNAs important for normal B-cell differentiation and B-cell-derived tumorigenesis [74-76].
Figure 2. Involvement of miRNAs on normal and malignant human B-cell differentiation.
The downregulation (↓) or upregulation (↑) of the miRNAs depicted here has been shown to be involved in the reaction to which they are adjacent. ALL: Acute Lymphoblastic Leukemia; CLL: Chronic Lymphocytic Leukemia; HL: Hodgkin Lymphoma; NHL: Non-Hodgkin Lymphoma; MM: Multiple Myeloma
DEVELOPMENT AND PROGRESSION OF CANCER
The development of cancer is a very complex process. During malignant expansion, cells acquire capabilities that allow tumor growth, such as sustained proliferative signaling, the ability to evade growth suppressors and resist cell death, replicative immortality, induced angiogenesis, activated invasion and metastasis, reprogrammed energy metabolism, and evasion from immune destruction. Tumor-promoting inflammation and genome instability and mutation also contribute to malignant proliferation [77, 78].
Due to their ability to control gene expression, deregulated miRNA expression is awarded a central role in oncogenesis. MiRNAs are recognized to function as oncogenes when their upregulation inhibits the expression of tumor suppressor genes, thereby favoring the development and progression of cancer by evading apoptotic and growth suppression mechanisms. Likewise, miRNAs can act as tumor suppressors, preventing the expression of proto-oncogenes and consequently hindering tumorigenesis, e.g., when inducing cell cycle arrest in malignant cells [79]. Their role is not always clear, as is the case of miR-29 in chronic lymphocytic leukemia (CLL) [80]. MiR-29 is downregulated in aggressive CLL causing the upregulation of TCL1, a critical oncogene, suggesting it may act as a tumor suppressor. However, it is involved in the pathogenesis of indolent CLL, where it is expressed at higher levels than in aggressive CLL and normal B-cells, which points to an oncogene function.
The mechanisms causing deregulated expression of miRNAs are not always known. However, Calin et al. [81, 82] showed in 2004 that they are frequently located in fragile sites and in cancer-associated genomic regions. More recent studies have documented epigenetic silencing of miRNAs by DNA methylation and chromatin modifications [83] as well as miRNA targeting of DNA methyltransferases [84, 85], histone deacetylases [86], and proteins involved in histone modifications [84, 85, 87] in multiple cancers. The impact of miRNA regulation of gene expression is illustrated in a more indirect manner when cancer cells truncate the 3′ UTR of target mRNAs by alternative polyadenylation, thus avoiding miRNA regulation [88, 89]. Here we present a number of studies based on either global miRNA expression profiling or functional in vitro/in vivo models that pinpoint the importance of the deregulated expression of miRNAs in the development and progression of hematological B-cell cancers.
miR-15 and miR-16
The first demonstration of the relevance of miRNAs in human cancer recognized the involvement of miR-15 and miR-16 in CLL. With the knowledge that deletions at chromosome 13q14 occur frequently in CLL, lymphomas, and solid cancers, Calin et al. [81] found that miR-15 and miR-16 are localized in a 30 kb genomic region at 13q14 also associated with translocations in CLL [7]. In CLL patients with a deletion of that region, both miRNAs are downregulated compared to normal CD5+ B-cells. The functional evaluation of their activity was provided in a mouse model where the minimal deleted region of 13q14, containing miR-15a/16-1, was conditionally deleted, resulting in CLL phenotype disease [90]. These results point to a tumor suppressor function for miR-15a and miR-16 in CLL (Table 1).
Table 1. MiRNA involvement in hematological malignancies.
| miRNA | Hematopoiesis | Ref | Development and progression of cancer | Suggested mechanism in hematological cancers | Ref | Response to treatment | Ref | Possible protein targets* |
|---|---|---|---|---|---|---|---|---|
| miR-15 miR-16 |
miR-16: regulates differentiation of late erythroid progenitor cells | [159] | Tumor-suppressor in CLL Possibly different roles in early and late-stage MM |
Cell cycle arrest Decrease in proliferation Antiangiogenic activity |
[7, 81, 91-93] | MiR-15a expression increased in MM cell lines in the presence of bortezomib, decreased when BMSC were added | [92] | Bcl2, cyclin D1, cyclin D2, CDC25A, pRb, AKT3, pAKT, rpS6, pERK, TAB3, VEGF miR-16: RARS |
| miR-17-92 cluster | Essential for Pro-B and Pre-B-cell survival | [74] | Oncogene in B-cell lymphoma miR-17: Tumor-suppressor in miR-92a-induced erythroleukemia |
Decrease in apoptosis (possibly due to elevated myc) Increases cell proliferation Promotes cell survival Promotes the development of stem cell properties or characteristics of early developmental lineages Activation of the PI3K/AKT pathway |
[103, 111, 113, 128, 160, 161] | Overexpression increased survival of MCL cell lines after treatment with topotecan, doxorubicin, or etoposide Downregulation suppressed tumor growth in a mouse model of MCL treated with doxorubicin miR-19a and miR-19b were downregulated in MM cell lines in the presence of ITF2357 miR-17: High expression associated with shorter OS in MM patients |
[128, 129, 142] | p53, Gata-1, PTEN, E2F1, Bim, PHLPP2 miR-17: Bcl2, STAT-5, Jak2, p21 miR-19a and miR-19b: SOCS1 miR-92: KLF2 [63] |
| miR-21 | --- | --- | Oncogene in MM and B and T-cell lymphoma | Decrease in apoptosis | [13, 99-104, 162] | Associated with resistance to CHOP in DLBCL | [130] | PTEN, PPCD4, Bim, SHIP1 |
| miR-27a | Promotes erythroid differentiation | [163] | --- | --- | --- | Lower expression in K562 cells resistant to doxorubicin | [134] | MDR1, STAT-5, GATA2 [159] |
| miR-28 | Involved in the GC reaction | [114] | Tumor-suppressor in GC-derived lymphomas | Decrease in proliferation and clonogenicity Increase in apoptosis |
[114] | --- | --- | BAG1, MAD2L1, RAP1B |
| miR-34a | Delays Pro-B to Pre-B-cell transition Essential for transdifferentiation from Pre-B-cells to macrophages |
[164, 165] | --- | --- | --- | In the presence of bortezomib, downregulation of miR-34a increased apoptosis of P493-6 cells miR-34a overexpression in TP53-mutated MM cell lines and mice inhibited tumor growth |
[125, 127] | Myc, p53, BCL2, CDK6, NOTCH1, Lef1, Foxp1 |
| miR-122 | --- | --- | --- | --- | --- | Protective action against treatment with bortezomib, MG132, and GSI-1 | [139] | cyclin G1 |
| miR-127-3p | --- | --- | --- | --- | --- | Prognostic role in MCL patients especially in combination with other clinical indicators | [141] | BLIMP1 |
| miR-146b-5p | --- | --- | Tumor-suppressor in DLBCL | Decrease in proliferation | [143] | Lower expression in CHOP-treated DLBCL patients with poorer OS | [143] | --- |
| miR-150 | Blocks Pro-B to Pre-B-cell transition High expression in T-cells Downregulation during erythroid and megakaryocytic differentiation |
[64, 66, 159] | Tumor-suppressor in NK/T-cell lymphoma | Increase in apoptosis (especially in the presence of inhibition of miR-21) Decrease in cell proliferation Control of aging and senescence by decreasing telomerase activity |
[2] | --- | --- | DKC1, AKT2, Myb, Survivin, Foxp1, cMYB |
| miR-155 | Controls GC reaction Regulates T helper cell differentiation |
[69] | Oncogene in B-cell lymphoma | Decrease in apoptosis | [109, 162] | --- | --- | PTEN, PPCD4, Bim, SHIP1, FXR1, Ago2, c-Maf, Pu.1, AID, CXCR4, JUN, GATA-3 [159] |
| miR-181a/b | Early differentiation of B-cells Decrease in proliferation of T-cells Decrease granulocytic and macrophage-like differentiation |
[62, 147, 164, 166] | Oncogenes in NHL and AML | NHL: Decrease in apoptosis AML: blockage of myeloid differentiation and infiltration of leukemic cells into the bone marrow and spleen |
[144, 145, 147, 148] | Promote apoptosis in fludarabine-exposed CLL cells that express p53 Prognostic potential in CLL showing downregulation during progressive disease |
[144, 145] | CXCR4 [63], BCL2, MCL1, TCL1, XIAP, PRKCD miR-181a: Bim, CAMKK1, CTDSPL miR-181b: AID |
| miR-221 miR-222 |
Downregulation is essential for expansion of erythroblasts | [115, 159] | Oncogenes in MM | Increase of DNA replication AKT activation |
[115, 116, 159] | --- | --- | p27Kip1, PTEN, p57Kip2, c-kit |
| miR-223 | Involved in the transition from GC cells to post-GC cells Controls granulocytic differentiation Essential for transdifferentiation from Pre-B-cells to macrophages | [36, 63, 165, 167] | --- | --- | --- | --- | --- | NFI-A, Lef1 |
| miR-320d | --- | --- | Tumor-suppressor in DLBCL | Decrease in proliferation | [143] | Lower expression in CHOP-treated DLBCL patients with poorer PFS and OS | [143] | --- |
| miR-331-5p | --- | --- | --- | --- | --- | Lower expression in K562 cells resistant to doxorubicin | [134] | MDR1 |
| miR-615-3p | --- | --- | --- | --- | --- | Prognostic role in MCL patients especially in combination with other clinical indicators | [141] | --- |
| miR-886-5p | --- | --- | --- | --- | --- | High expression associated with shorter OS in MM patients | [142] | NR3C1 |
Based on references mentioned for Hematopoiesis, Development and progression of cancer, and Response to treatment, unless otherwise stated.
Ago2: Argonaute-2; AID: Activation-induced cytidine deaminase; AKT3: AKT serine/threonine protein kinase-3; BAG1: Bcl2-associated athanogene; DKC1: Dyskerin; FKBP51: FK506-binding protein 51; FXR1: Fragile-X mental retardationrelated protein 1; MAD2L1: MAD2 mitotic arrest deficient-like 1; PDCD4: Programmed cell death 4; PHLPP2: PH domain and Leucine-rich repeat Protein Phosphatase 2; PTEN: Phosphatase and tensin homologue; RAP1B: member of RAS oncogene family; RARS: Arginyl-tRNA synthetase gene; rpS6: ribosomal-protein-S6; SHIP1: Src homology-2 domain-containing inositol 5-phosphatase 1; SOCS1: Suppressor of cytokine signaling 1; TAB3: TGF-beta activated kinase 1/MAP3K7 binding protein 3; VEGF: Vascular endothelial growth factor; XIAP: X-linked inhibitor of apoptosis protein.
Similar results have been found in multiple myeloma (MM). Roccaro et al. [91] identified a miRNA signature for relapsed/refractory MM patients that included downregulation of both miR-15a and miR-16. Additionally, 9 MM patients with chromosome 13 deletion were devoid of expression of these two miRNAs, while 6 MM patients without the deletion presented decreased expression compared to bone marrow from 4 healthy donors. Transfection of MM cell lines with miR-15a and miR-16 simultaneously [91] or with miR-15a alone [92] caused cell cycle arrest and decreased proliferation, supporting a tumor suppressor role. Both studies found an indirect regulation of the vascular endothelial growth factor (VEGF) mature protein, crucial in the pathogenesis and progression of MM, which presented decreased levels after transfection with the miRNAs.
The function of miR-15a in MM has not been completely elucidated since its expression in 52 newly diagnosed patients was found to be higher than in 2 healthy donors [93]. This suggests a different role for miR-15a in primary and advanced MM, illustrating that the function of a given miRNA may depend on cellular context.
miR-21
MiR-21 has been reported to be upregulated and to possess oncogenic activity in several cancers, including breast cancer [94], colon adenocarcinoma [3], glioblastoma [95], non-small cell lung cancer [96], esophageal squamous cell carcinoma [97], and melanoma [98]. Overall, miR-21 has been shown to be involved in the development and progression of cancer, with higher expression in more advanced stages of the disease. Although the studies on hematological conditions are limited in number in comparison with solid tumors, miR-21 has also been found to be upregulated in MM, B- and T-cell lymphoma patients and cancer cell lines, including DLBCL [13, 99-103]. Additionally, Medina and colleagues [104] showed that transgenic mice overexpressing miR-21 developed signs of hematological malignancies, which in the majority of the mice resulted in lymphomas of the B-cell lineage. When the overexpression of miR-21 was blocked with doxycycline tumor regression was observed, allowing the authors to suggest oncomiR-21 addiction, with miR-21 impacting not only the initiation of a malignant process but also its development and maintenance, making it an important potential therapeutic target.
Bone marrow stromal cells (BMSC) have been shown to exert a protective effect on MM cells, contributing to their proliferation. Recently, miR-21 was found to be upregulated in MM cells grown with BMSC, with levels even higher than in MM patients. This was observed both in primary patient samples and in the INA-6 cell line compared to normal B-cells [105]. The inhibition of miR-21 with oligonucleotides contributed to an anti-tumor effect in vitro in cells with high basal expression and in vivo in mouse bearing xenografts of high expression human cells [105]. This suggests that downregulation of miR-21 could potentially be a therapeutic strategy, but not using a “one size fits all” approach.
miR-155
GC B-cells express high levels of miR-155, while relatively low expression is observed in pre and post-GC B-cells, indicating a role in the GC reaction [57, 63, 67, 68]. Upregulated miR-155 expression compared to normal counterpart B-cells or tissues was observed for several hematological cancers, including DLBCL of the most aggressive type (ABC-DLBCL) [13, 106, 107], PMBCL, and Hodgkin Lymphoma (HL) [108]. Recently, epigenetically reactivated miR-155 was documented in CLL, where the promoter of miR-155 exhibited a significant decrease in local DNA methylation accompanied by consistent transcriptional upregulation of the mature miR-155 [83]. Functionally, there is evidence of oncogenic properties for miR-155 because its overexpression in transgenic mice consistently caused B-cell lymphoproliferative disease [109].
miR-17-92 cluster
The miR-17-92 cluster is located on chromosome 13q31, a region frequently amplified in different tumor types [110]. It encodes 6 miRNAs: miR-17, miR-18a, miR-19a, miR-19b-1, miR-20a, and miR-92-1. It is highly expressed in progenitor B-cells, and expression diminishes as cells mature. The miR-17-92 cluster antagonizes the expression of BIM, a pro-apoptotic protein, thereby favoring the survival of B-cell progenitors [74]. This cluster, as well as individual miRNAs from the cluster, was found to be upregulated in samples from DLBCL, mantle cell lymphoma (MCL), and acute lymphocytic leukemia (ALL) [74, 111, 112]. In a functional in vivo murine model of human B-cell lymphoma overexpressing the c-myc oncogene, mice with simultaneous overexpression of a miR-17-19b-1 truncated cluster experienced accelerated disease development, which culminated in highly malignant B-cell lymphoma with lower apoptotic capacity and overall survival (OS) [111]. These results suggest that the miR-17-92 cluster has oncogenic properties. However, the function of the individual miRNAs may be dependent on factors such as the lineage of the cells. For example, Li et al. [113] recently reported that miR-17 was able to delay the development of miR-92a-induced erythroleukemia.
miR-28
MiR-28 has been shown to be downregulated in GC-derived lymphomas compared to GC B-cells [114]. The authors provided functional evidence of the role of this miRNA in the proliferation of malignant cells, finding reduced proliferation and clonogenicity, increased apoptosis, and G1-arrest when miR-28 was overexpressed. This suggests miR-28 may have a tumor suppressor function. However, these results need further validation.
miR-221 and miR-222
Deregulated expression of miR-221 and miR-222 has also been associated with tumor development. Although able to inhibit erythroleukemic cell growth through impairment of erythroid differentiation and proliferation [115], they are reported to possess oncogenic activity. Di Martino and colleagues [116] recently demonstrated their potential as therapeutic targets by showing antitumor activity from miR-221 and miR-222 inhibitors in two MM cell lines expressing moderate or high levels of both miRNAs. The inhibitors were also tested on an MM mouse model where the miR-221 inhibitor, able to downregulate both miRNAs, presented higher activity than the miR-222 inhibitor alone or the two used in combination. This is one of several examples of the possible therapeutic application of the modulation of the expression of miRNAs.
RESPONSE TO TREATMENT
One of the main reasons for the dismal OS of patients suffering from different types of B-cell-derived cancer is treatment resistance. There are two clinically distinct types of resistance: intrinsic and acquired. The former causes primary refractory disease, reflecting characteristics within the tumor at time of diagnosis that determine the ability of the tumor cells to thwart the effectiveness of a given drug. In contrast, acquired resistance occurs in initially responsive patients, and it is caused by a selection process where chemoresistant clones overgrow the sensitive ones [117-119]. Early detection of resistance is of great importance. Therefore, there is a strong need for biomarkers that ensure a more accurate classification of B-cell-derived malignancies or that are able to guide selection of alternative treatments. Defining miRNA involvement in resistance will be useful to guide treatment selection and help monitor treatment administration. It could provide information on the mechanisms activated by the drug and make miRNAs themselves biomarkers of both intrinsic and acquired chemoresistance, thereby improving primary treatment choice and circumventing intrinsic resistance.
MiRNAs can act as prognostic and predictive biomarkers. The prognostic or predictive role can be explained by the anticipation of how a patient is expected to respond to a combined intervention or to one drug in particular, respectively, based on the presence or absence of a miRNA. Understanding the involvement and the possible use of miRNAs in response to treatment is invaluable to tailor the therapeutic approach to each individual patient. In the following two sections we will present the potential of miRNAs in guiding treatment strategy. We first present studies that show how the in vitro or in vivo manipulation of the expression of specific miRNAs can affect the response to a given drug, and consequently unravel their biological roles in treatment response by pinpointing the potential of miRNAs as predictive biomarkers. In the second section we will focus on miRNAs that have demonstrated a correlation between expression levels and outcome of treatment, illustrating the role of miRNAs as prognostic biomarkers.
MiRNAs as predictive biomarkers of treatment response
The knowledge of how a patient is expected to respond to a specific drug or regimen is very helpful in regard to deciding the best possible treatment. It can prevent the occurrence of toxicity or sub-therapeutic dosages, or indicate that another therapeutic regimen may be more suitable for the patient in question. This information is provided by predictive biomarkers. Understanding the biological function of miRNAs may suggest that a specific drug is effective or not. However, the study of predictive biomarkers, which provide limited biological insight, is extremely important as it can more strongly support the development of individualized treatment strategies.
miR-15 and miR-16
MiR-15 and miR-16 have been linked to sensitivity to chemotherapeutic agents. Hao et al. [92] showed that MM U266 and H929 cell lines were sensitive to bortezomib and the expression of miR-15a was increased in the presence of the drug. When the bortezomib-treated cells were allowed to interact with BMSC the tumor suppressor effect was decreased along with miR-15a levels. The expression of the anti-apoptotic protein Bcl-2, a target of miR-15a, was upregulated in these cells contributing to the protection of MM cells against bortezomib-induced apoptosis and promoting their survival. As mentioned earlier, transfection with miR-15a and miR-16 promotes cell cycle arrest and inhibits proliferation of MM cell lines [91, 92]. Therefore, induction of these miRNAs may be one of the mechanisms of action of bortezomib. These studies suggest that overexpression of miR-15a and miR-16 can help overcome resistance towards bortezomib in MM.
miR-34a
The role of miR-34a in cancer has not been completely clarified. Although there are publications suggesting a tumor-favoring activity, the majority of studies point to a tumor suppressor function. Hence, activity most likely depends on the cellular and genetic microenvironment [120, 121]. The miR-34a gene is located on chromosome 1p36.22, a region frequently altered in cancer [82]. Its promoter can become hypermethylated, causing reduced levels of miR-34a [122]. Downregulated miR-34a expression is observed in gastric marginal zone B-cell lymphoma of MALT type that transforms into gastric DLBCL [123]. Reestablishing miR-34a expression in a lymphoma xenograft model reduced tumor growth by 76%, suggesting a true therapeutic utility of this miRNA [124]. Accordingly, miR-34a overexpression in TP53-mutated MM cell lines and in mice bearing xenografts of these cells showed tumor growth inhibition and even regression, supporting a tumor suppressor role when TP53 is mutated [125]. However, the genetic and deregulated expression background of the B-cell tumors may dramatically affect the function of miR-34a. On one hand, miR-34a is able to mitigate resistance to taxol and platinum-based agents in solid tumors [126]. On the other hand, Sotillo et al. [127] showed that transfection of miR-34a into a B-cell line genetically similar to Burkitt's Lymphoma, overexpressing Myc, downregulated Myc and p53 protein levels, which led to decreased apoptosis. In the presence of bortezomib, upregulation of miR-34a inhibited p53-dependent drug-induced apoptosis. MiR-34a downregulation increased the apoptotic activity, illustrating that in tumors with deregulated Myc expression miR-34a confers drug resistance and its downregulation could be a strategy to increase response to bortezomib. Thus, the initial mutational status and subsequent treatment-induced clonal selection of malignant cells affect the intrinsic and acquired drug resistance, respectively, and also the miRNAs involved in the drug response.
miR-17-92 cluster
Treatment outcome of MCL patients may also be influenced by miRNA expression. MiRNAs of the cluster miR-17-92 were shown to be upregulated in these patients, with 2 to 3-fold greater levels than in normal naïve B-cells. High levels of C13orf25, the gene that encodes the cluster, were correlated with poorer OS [128]. Overexpression of the cluster by stable transduction in MCL Z138c and Granta-519 cell lines increased cell survival after exposure to topotecan, doxorubicin, or etoposide, while knockdown of miR-17-92 was shown to significantly suppress tumor growth in a mouse model of MCL treated with doxorubicin. These miRNAs also impacted the response to treatment of other cells. In the presence of the histone deacetylase ITF2357, a drug that induces apoptosis in MM cells, miR-19a and miR-19b were 25-57% downregulated in KMS18 and KMS12 cells [129]. This was reported as a possible mechanism of action of the drug. Downregulation of miR-17-92 might therefore be beneficial to reduce treatment resistance in MCL.
miR-21
MiR-21 was recently suggested to play a part in resistance to the CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone) regimen in DLBCL, as its knockdown in CRL2631 cells increased the sensitivity to treatment via phosphatase and tensin homolog (PTEN) upregulation [130]. This is in agreement with observations in MM, where miR-21 inhibition in combination with dexamethasone, doxorubicin, or bortezomib inhibited myeloma cell survival more effectively than any treatment alone [131]. Together with the findings of upregulation of miR-21 in DLBCL, MM, and several other cancers, these results support its oncogenic role and potential as a therapeutic target. Simultaneously, diminished expression of miR-21 might be predictive of tumors likely to be responsive to CHOP, dexamethasone, doxorubicin, or bortezomib.
miR-155
Epstein-Barr virus (EBV)-associated DLBCL is a rare disease with poor survival due to resistance to chemotherapy [132]. EBV expressing latent membrane protein-1 (LMP1) activates the Akt pathway by phosphorylation and upregulates the antiapoptotic MCL1 through miR-155 expression, which contributes to the survival of the rituximab-treated cells. This shows the involvement of miR-155 in resistance to rituximab. Knockdown of miR-155 expression is able to reduce Akt phosphorylation, resulting in the significantly increased death of EBV-positive cells treated with rituximab [133]. These results indicate that inhibition of miR-155 could be a valuable approach for the treatment of EBV-associated B-cell lymphomas.
miR-331-5p and miR-27a
Three myelogenous leukemia cell lines derived from K562 cells with gradually increasing resistance to doxorubicin showed an inverse correlation between the expression of two miRNAs, miR-331-5p and miR-27a, and the degree of resistance to the drug [134]. When transfected with these miRNAs, the doxorubicin-resistant cells exhibited a higher sensitivity to the drug and decreased expression of P-glycoprotein (P-gp). This could be due to the targeting of the multiple drug resistance protein 1 (MDR1) gene, which encodes for P-gp and whose increase is identified as a possible mechanism of resistance. In fact, expression of MDR1 can be increased in B-cell lymphoma Daudi cells by treatment with doxorubicin, which is an example of the development of acquired resistance [135]. Additionally, Raji cells previously co-cultured with follicular dendritic cells presented higher MDR1 expression and a lower apoptotic rate after exposure to doxorubicin compared to Raji cells cultured alone [136]. Feng et al. [134] also found that relapsing leukemia patients had lower expression of miR-27a and miR-331-5p than patients at diagnosis or in complete remission, supporting the tumor suppressor role of these miRNAs and their possible application in surpassing resistance to doxorubicin.
Predictive miRNAs in therapeutic intervention
The predictive potential of miRNAs is directly linked to their ability to be used in the treatment of malignancies. MiRNAs shown to contribute to resistance to a particular drug or regimen are potential targets for treatment, thus, obvious candidates for drug discovery, while miRNAs that directly or indirectly increase the response to treatment have therapeutic potential on their own. The latter are plausible candidates for studies of drug formulation. Therefore, the study of predictive biomarkers provides information on how to guide the treatment of patients with the options currently available and also on possible future therapeutic strategies.
miR-122
MiR-122 is so far the only miRNA that has been directly targeted in a clinical trial. It is involved in the cholesterol and lipid metabolism and also in hepatitis C virus replication [137]. Antagonization of miR-122 is being evaluated for the treatment of hepatitis C virus infection in human clinical trials [138]. Initially thought to be liver-specific, miR-122 was later shown to be expressed in cutaneous T-cell lymphoma (CTCL) cell lines [139]. Its overexpression in these cells exerted a protective action against chemotherapy-induced apoptosis, namely against bortezomib, MG132 (proteasome inhibitors), and GSI-1 (a γ-secretase inhibitor). The exact mechanism is unknown; however, Akt and p53 seem to be involved. The results suggest that inhibiting miR-122 may be a strategy to improve treatment outcome in CTCL patients [139].
MiRNAs as prognostic biomarkers of treatment response
The potential of miRNAs as prognostic biomarkers under homogeneous treatment, even when their biological roles are not validated, should not be disregarded as this has a direct utility in clinical practice. The anticipation of how a disease will develop is of tremendous importance as it can make a difference in the choice of the treatment course, chemotherapy, or palliative care. It is, understandably, a question patients would like to have answered not only at time of diagnosis but throughout the progression and treatment of the disease. Research for prognostic tools is essential for diseases like cancer that have such a dramatic impact on quality of life and survival. To the best of our knowledge, no single miRNA or panel of miRNAs is at present implemented as validated biomarkers of prognosis in a routine clinical setting, not even when the translation potential is suggested as in the studies mentioned below.
The study of prognostic biomarkers, which offer less biological insight, may initially seem less relevant than studies of predictive biomarkers. However, by showing a correlation with clinical outcome, prognostic biomarkers demonstrate potential utility in a clinical setting and their use is reliable even without fully understanding the mechanism by which they function.
miR-222
After identifying a miRNA signature in cell lines able to distinguish ABC from GCB DLBCL, Alencar and colleagues [10, 11] set out to find which miRNAs of the signature correlated with survival in DLBCL patients. In a cohort of 176 patients treated with R-CHOP (rituximab added to the CHOP regimen), they found that increased miR-18a was associated with shorter OS, high miR-222 correlated with shorter progression-free survival (PFS), and increased miR-181a was positively associated with PFS. Shortly afterwards, Montes-Moreno et al. [12] published their findings on a prognostic miRNA signature found in a 36-patient discovery cohort and tested in 240 patients. It comprised 9 miRNAs and was able to predict poor clinical outcome in patients treated with the same regimen as in the Alencar et al. study. MiR-222 was present in both signatures, reinforcing its potential as a prognostic biomarker.
miR-34a
MiR-34a has also been correlated with clinical outcome. It was found to be downregulated in DLBCL samples. Although the study did not find an association between miR-34a and OS, the combination of low miR-34a and high FOXP1, BCL2, and p53, validated targets of miR-34a [126], was associated with poorer OS in a multivariate survival analysis [140]. The lack of association with miR-34a alone could be due to the different therapeutic approaches used among patients. This study suggests that several factors are involved in the activity of miR-34a, reinforcing the relevance of the characteristics of the tumor and its surroundings on the activity of this miRNA and supporting a classification of diseases based on its COO.
miR-127-3p, miR-615-3p, miR-17, and miR-886-5p
Using a training set of NHL, Goswami and colleagues [141] identified an 11-miRNA signature that distinguished indolent from aggressive forms of lymphomas, highlighting the diagnostic potential of miRNAs. Two of these, miR-127-3p and miR-615-3p, seemed able to provide information on the prognosis of MCL patients, especially if used in combination with the current clinical indicators Ki-67, a marker of cell proliferation, used with a redefined cut-off, or the MCL International Prognostic Index. Wu et al. [142] identified a classifier comprising 2 miRNAs, miR-17 and miR-886-5p, that distinguished three subgroups of MM patients with different OS, improving the classification provided by the International Staging System and Fluorescence In-Situ Hybridization-based abnormalities. Therefore, the combination of miRNA expression with already established prognostic biomarkers is promising for clinical use.
miR-146b-5p and miR-320d
A recent report has shown that miR-146b-5p and miR-320d have potential to discriminate between CHOP-treated DLBCL patients with different clinical outcomes [143]. Low expression of both miRNAs compared to the median was found in patients with poorer PFS and/or OS. In addition, miR-320d was more highly expressed in GCB-DLBCL patients.
miR-181a/b
MiR-181a and miR-181b also hold promise as prognostic biomarkers. MiR-181b has been shown downregulated in CLL patients and its expression further decreases with progression of the disease [144, 145]. Both miRNAs target anti-apoptotic proteins in these cells and induce apoptosis in primary cell cultures where p53 is normally expressed [145]. In a meta-analysis performed recently, downregulated expression of miR-181a/b was correlated with poor OS in hematological malignancies [146]. However, this could be another situation where the microenvironment deeply affects the activity of miRNAs, as inhibition of expression of the miR-181 family improved the condition of acute myeloid leukemia (AML) mice models and high miR-181a was suggested to behave as an oncogene also in NHL cell lines and primary lymphoma cells [147, 148].
Dicer
The impact of deregulated expression of Dicer, essential for the biogenesis of miRNAs, has also been examined. Zhou et al. [93] found that its knockdown in one myeloma cell line led to cell cycle arrest and increased apoptotic events, and Adams and colleagues [149] showed that B-cell lymphomas require Dicer for survival. However, Sarasquete et al. [150] showed an association between its upregulation and longer PFS in symptomatic MM patients. Hence, the effect of changes in the expression of this endoribonuclease on prognosis is not yet clear. This is supported by a recently published review that assessed the prognostic significance of its expression in different types of tumor, showing that different results have been found not only between distinct diseases but also among studies focusing on the same disease [151]. It is necessary to conduct large studies to make statements on how Dicer, and other catalyzers of miRNA biogenesis, may impact or predict the development of diseases.
PERSPECTIVES
MiRNAs are molecules of the utmost biological importance, making them interesting as diagnostic, prognostic, and predictive biomarkers. In this review, we have presented examples of how they could be useful in clinical practice. Certain miRNAs, specifically miR-15, miR-21, and the miR-17-92 cluster, have been associated with the development or progression of hematological malignancies and response to treatment. The exciting potential of these miRNAs suggests that more clinical studies should focus on them so that their measurement can have clinical utility in the years to come.
However, we have also shown that the results are sometimes contradictory. This might be explained by several factors, including the degree of cellular differentiation or the progression of disease at the moment of sample collection, the sampling material and its quality, the number of samples analyzed, the cellular context, scientific approaches, and statistical analyses. Another factor that may impact the different results, or the lack of associations in many studies, is the fact that miRNAs are usually expressed at low levels. A small fold change may be sufficient to impact the expression of their targets and the networks they interact with but may not be easily detected.
Instead of tumor biopsies, serum and plasma are sometimes used for miRNA analysis. One example is the high expression of 3 miRNAs (miR-494, miR-1973, and miR-21) observed in plasma from classical HL patients before treatment, decreasing to levels similar to healthy individuals at remission [152]. Sevcikova et al. [153] found that miR-29a, normalized against miR-16, was differentially expressed between serum from 91 MM patients and 30 healthy donors, although no association was identified with disease stage. A five-miRNA serum signature with predictive potential towards R-CHOP treatment in DLBCL patients has also been recently reported [154]. There is not always a clear correlation between miRNA expression in these fluids and tumor tissue [152], which according to some authors may be because a tumor would have to develop for several years before the expression of circulating miRNAs could be used as a tumor biomarker [155]. There can also be differences in measurements between serum and plasma, which can be due to the method of extraction of miRNAs. The presence of cellular components and hemolysis affects miRNAs differently in blood fluids [156], highlighting the importance of improvements in the technique. However, the stable expression of miRNAs in serum and plasma and their easy access in these biofluids make their potential as biomarkers evident. This is also supported by the recent study by Chen W et al. [157], who found a linear correlation between miR-21 expression in serum and tumor tissue in 30 DLBCL patients. Additionally, based on serum from 62 patients, they found higher miR-21 levels in patients with a better prognosis.
MiRNA isolation and measurement with PCR in bone marrow smears has been recently reported [158]. The authors found differentially expressed miRNAs between 3 normal donors and 5 FL patients. This type of sample presents the advantage of being easily stored and transported at room temperature. These results suggest that bone marrow smears could be another option for screening miRNA expression.
As shown above, miRNAs are involved in many cellular processes. They are essential in the differentiation, maturation, and metabolism of cells, capable of controlling gene expression and the expression of other non-coding RNAs. This pervasive activity leads us to hypothesize that they also play a role in defining the COO of B-cell malignancies and drug-specific resistance. We hypothesize that the characterization of B-cell subsets will help classify B-cell malignancies and predict their response to treatment. To test this, we are characterizing primary B-cell subsets purified from various lymphatic tissues. Additionally, we have performed in vitro drug screens in DLBCL and MM cell lines and scrutinized their mRNA and miRNA expression to identify drug response profiles. Tracing the COO of tumors will allow a better insight into the cancer biology and a more thorough characterization of the tumor of each patient. The combination of this knowledge with associations between expression of specific miRNAs and drug response will enable the prediction of the most beneficial treatment for each patient.
Although the role of specific miRNAs in the development of cancer, response to treatment, and development of resistance still needs further clarification, their potential in therapeutic intervention and as prognostic and predictive biomarkers is undeniable. It is our hope that future studies on miRNAs will provide support for the assignment of individual patients to specific drug response profiles and the consequent development of personalized treatment strategies.
REFERENCES
- 1.Jansson MD, Lund AH. MicroRNA and cancer. Mol. Oncol. 2012;6(6):590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watanabe A, Tagawa H, Yamashita J, Teshima K, Nara M, Iwamoto K, Kume M, Kameoka Y, Takahashi N, Nakagawa T, Shimizu N, Sawada K. The role of microRNA-150 as a tumor suppressor in malignant lymphoma. Leukemia. 2011;25(8):1324–34. doi: 10.1038/leu.2011.81. [DOI] [PubMed] [Google Scholar]
- 3.Schetter A, Leung S, Sohn J, Zanetti K, Bowman E, Yanaihara N, Yuen S, Chan T, Kwong D, AU G, Liu C, Calin G, Croce C, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. 2000;403(6769):503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 5.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002;346(25):1937–47. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 6.Lenz G, Staudt LM. Aggressive lymphomas. N. Engl. J. Med. 2010;362(15):1417–29. doi: 10.1056/NEJMra0807082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shaffer AL, Young RM, Staudt LM. Pathogenesis of human B cell lymphomas. Annu. Rev. Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campo E, Swerdlow SH, Harris NL, Pileri S, Stein H, Jaffe ES. The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood. 2011;117(19):5019–32. doi: 10.1182/blood-2011-01-293050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swerdlow S, Campo E, Harris N, Jaffe E, Pileri S, Stein H, Thiele J, Vardiman J, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer (IARC); 2008. [Google Scholar]
- 10.Alencar AJ, Malumbres R, Kozloski G a, Advani R, Talreja N, Chinichian S, Briones J, Natkunam Y, Sehn LH, Gascoyne RD, Tibshirani R, Lossos IS. MicroRNAs Are Independent Predictors of Outcome in Diffuse Large B-Cell Lymphoma Patients Treated with R-CHOP. Clin. Cancer Res. 2011;17(12):4125–4135. doi: 10.1158/1078-0432.CCR-11-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malumbres R, Sarosiek K a, Cubedo E, Ruiz JW, Jiang X, Gascoyne RD, Tibshirani R, Lossos IS. Differentiation stage-specific expression of microRNAs in B lymphocytes and diffuse large B-cell lymphomas. Blood. 2009;113(16):3754–64. doi: 10.1182/blood-2008-10-184077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montes-Moreno S, Martinez N, Sanchez-Espiridión B, Díaz Uriarte R, Rodriguez ME, Saez A, Montalbán C, Gomez G, Pisano DG, García JF, Conde E, Gonzalez-Barca E, Lopez A, et al. miRNA expression in diffuse large B-cell lymphoma treated with chemoimmunotherapy. Blood. 2011;118(4):1034–40. doi: 10.1182/blood-2010-11-321554. [DOI] [PubMed] [Google Scholar]
- 13.Lawrie CH, Soneji S, Marafioti T, Cooper CDO, Palazzo S, Paterson JC, Cattan H, Enver T, Mager R, Boultwood J, Wainscoat JS, Hatton CSR. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int. J. Cancer. 2007;121(5):1156–61. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 14.Pasqualucci L. The genetic basis of diffuse large B-cell lymphoma. Curr. Opin. Hematol. 2013;20(4):336–44. doi: 10.1097/MOH.0b013e3283623d7f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnsen HE, Bergkvist KS, Schmitz A, Kjeldsen MK, Hansen SM, Gaihede M, Nørgaard MA, Bæch J, Grønholdt M-L, Jensen FS, Johansen P, Bødker JS, Bøgsted M, et al. Cell of origin associated classification of B-cell malignancies by gene signatures of the normal B-cell hierarchy. Leuk. Lymphoma. 2014;55(6):1251–60. doi: 10.3109/10428194.2013.839785. [DOI] [PubMed] [Google Scholar]
- 16.Laursen MB, Falgreen S, Bødker JS, Schmitz A, Kjeldsen MK, Sørensen S, Madsen J, El-Galaly TC, Bøgsted M, Johnsen HE, (MSCNET) MSCN Human B-cell cancer cell lines as a preclinical model for studies of drug effect in diffuse large B-cell lymphoma and multiple myeloma. Exp. Hematol. 2014 doi: 10.1016/j.exphem.2014.07.263. [DOI] [PubMed] [Google Scholar]
- 17.Cai X, Hagedorn C, Cullen B. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425(6956):415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 19.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein E, Caudy a a, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409(6818):363–6. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 21.Hutvágner G, McLachlan J, Pasquinelli a E, Bálint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293(5531):834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 22.Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106(1):23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- 23.Kok KH, Ng M-HJ, Ching Y-P, Jin D-Y. Human TRBP and PACT directly interact with each other and associate with dicer to facilitate the production of small interfering RNA. J. Biol. Chem. 2007;282(24):17649–57. doi: 10.1074/jbc.M611768200. [DOI] [PubMed] [Google Scholar]
- 24.Iorio M V, Croce CM. microRNA involvement in human cancer. Carcinogenesis. 2012;33(6):1126–33. doi: 10.1093/carcin/bgs140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sibley CR, Seow Y, Saayman S, Dijkstra KK, El Andaloussi S, Weinberg MS, Wood MJ a. The biogenesis and characterization of mammalian microRNAs of mirtron origin. Nucleic Acids Res. 2012;40(1):438–48. doi: 10.1093/nar/gkr722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, You X, Chen T, Mackowiak SD, Friedländer MR, Weigt M, Du H, Gogol-Döring A, Chang Z, Dieterich C, Hu Y, Chen W. Global profiling of miRNAs and the hairpin precursors: insights into miRNA processing and novel miRNA discovery. Nucleic Acids Res. 2013;41(6):3619–34. doi: 10.1093/nar/gkt072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 28.Zhao G, Yu D, Weiss MJ. MicroRNAs in erythropoiesis. Curr. Opin. Hematol. 2010;17(3):155–62. doi: 10.1097/MOH.0b013e328337ba6c. [DOI] [PubMed] [Google Scholar]
- 29.Bhayani M, Calin G, Lai S. Functional relevance of miRNA* sequences in human disease. Mutat. Res. 2012;731(1-2):14–9. doi: 10.1016/j.mrfmmm.2011.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ro S, Park C, Young D, Sanders KM, Yan W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007;35(17):5944–53. doi: 10.1093/nar/gkm641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagos-Quintana M, Rauhut R, Meyer J, Borkhardt A, Tuschl T. New microRNAs from mouse and human. RNA. 2003;9(2):175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamura K, Phillips M, Tyler D, Duan H, Chou Y, Lai E. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008;15(4):354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuchenbauer F, Mah SM, Heuser M, McPherson A, Rüschmann J, Rouhi A, Berg T, Bullinger L, Argiropoulos B, Morin RD, Lai D, Starczynowski DT, Karsan A, et al. Comprehensive analysis of mammalian miRNA* species and their role in myeloid cells. Blood. 2011;118(12):3350–8. doi: 10.1182/blood-2010-10-312454. [DOI] [PubMed] [Google Scholar]
- 34.Pichiorri F, De Luca L, Aqeilan RI. MicroRNAs: New Players in Multiple Myeloma. Front. Genet. 2011;2(May):22. doi: 10.3389/fgene.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung AKL, Sharp PA. Quantifying Argonaute proteins in and out of GW/P-bodies: implications in microRNA activities. Adv. Exp. Med. Biol. 2013;768:165–182. doi: 10.1007/978-1-4614-5107-5_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N, Mancini M, Nanni M, Cimino G, et al. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119(17):4034–46. doi: 10.1182/blood-2011-08-371344. [DOI] [PubMed] [Google Scholar]
- 37.Tan Y, Zhang B, Wu T, Skogerbø G, Zhu X, Guo X, He S, Chen R. Transcriptional inhibiton of Hoxd4 expression by miRNA-10a in human breast cancer cells. BMC Mol. Biol. 2009;10(12):12. doi: 10.1186/1471-2199-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Younger ST, Corey DR. Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucleic Acids Res. 2011;39(13):5682–91. doi: 10.1093/nar/gkr155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moretti F, Thermann R, Hentze MW. Mechanism of translational regulation by miR-2 from sites in the 5′ untranslated region or the open reading frame. RNA. 2010;16(12):2493–502. doi: 10.1261/rna.2384610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandke P, Wyatt N, Fraser J, Bates B, Berberich SJ, Markey MP. MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One. 2012;7(8):e42034. doi: 10.1371/journal.pone.0042034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schnall-Levin M, Rissland OS, Johnston WK, Perrimon N, Bartel DP, Berger B. Unusually effective microRNA targeting within repeat-rich coding regions of mammalian mRNAs. Genome Res. 2011;21(9):1395–403. doi: 10.1101/gr.121210.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimakami T, Yamane D, Jangra RK, Kempf BJ, Spaniel C, Barton DJ, Lemon SM. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U. S. A. 2012;109(3):941–6. doi: 10.1073/pnas.1112263109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts APE, Lewis AP, Jopling CL. miR-122 activates hepatitis C virus translation by a specialized mechanism requiring particular RNA components. Nucleic Acids Res. 2011;39(17):7716–29. doi: 10.1093/nar/gkr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truesdell SS, Mortensen RD, Seo M, Schroeder JC, Lee JH, LeTonqueze O, Vasudevan S. MicroRNA-mediated mRNA translation activation in quiescent cells and oocytes involves recruitment of a nuclear microRNP. Sci. Rep. 2012;2:842. doi: 10.1038/srep00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30(4):460–71. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nat. Rev. Immunol. 2006;6(2):107–16. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- 48.Pelanda R, Torres RM. Central B-cell tolerance: where selection begins. Cold Spring Harb. Perspect. Biol. 2012;4(4):a007146. doi: 10.1101/cshperspect.a007146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perez-Andres M, Paiva B, Nieto WG, Caraux a, Schmitz a, Almeida J, Vogt RF, Marti GE, Rawstron a C, Van Zelm MC, Van Dongen JJM, Johnsen HE, Klein B, et al. Human peripheral blood B-cell compartments: a crossroad in B-cell traffic. Cytometry B. Clin. Cytom. 2010;78(Suppl 1(May)):S47–60. doi: 10.1002/cyto.b.20547. [DOI] [PubMed] [Google Scholar]
- 50.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu. Rev. Immunol. 2005;23(9):487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 51.Xu Z, Fulop Z, Zhong Y, Evinger AJ, Zan H, Casali P. DNA lesions and repair in immunoglobulin class switch recombination and somatic hypermutation. Ann. N. Y. Acad. Sci. 2005;1050:146–62. doi: 10.1196/annals.1313.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Allen CDC, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27(2):190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vasilatou D, Papageorgiou S, Pappa V, Papageorgiou E, Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. Eur. J. Haematol. 2010;84(1):1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Sankaran VG, Lodish HF. MicroRNAs in erythroid and megakaryocytic differentiation and megakaryocyte-erythroid progenitor lineage commitment. Leukemia. 2012;26(11):2310–6. doi: 10.1038/leu.2012.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koralov SB, Muljo S a, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132(5):860–74. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 56.Di Lisio L, Martinez N, Montes-Moreno S, Piris-Villaespesa M, Sanchez-Beato M, Piris M a. The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood. 2012;120(9):1782–90. doi: 10.1182/blood-2012-05-402784. [DOI] [PubMed] [Google Scholar]
- 57.Jensen K, Brusletto BS, Aass HCD, Olstad OK, Kierulf P, Gautvik KM. Transcriptional profiling of mRNAs and microRNAs in human bone marrow precursor B cells identifies subset- and age-specific variations. PLoS One. 2013;8(7):e70721. doi: 10.1371/journal.pone.0070721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang J, Jima DD, Jacobs C, Fischer R, Gottwein E, Huang G, Lugar PL, Lagoo AS, Rizzieri D a, Friedman DR, Weinberg JB, Lipsky PE, Dave SS. Patterns of microRNA expression characterize stages of human B-cell differentiation. Blood. 2009;113(19):4586–94. doi: 10.1182/blood-2008-09-178186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martinez J, Busslinger M. Life beyond cleavage: the case of Ago2 and hematopoiesis. Genes Dev. 2007;21(16):1983–8. doi: 10.1101/gad.1591407. [DOI] [PubMed] [Google Scholar]
- 60.Xu S, Guo K, Zeng Q, Huo J, Lam K-P. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119(3):767–76. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 61.Belver L, Yébenes VG, De Ramiro AR. microRNAs prevent the generation of autoreactive antibodies. 2010;33(5):713–722. doi: 10.1016/j.immuni.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. (80-) [DOI] [PubMed] [Google Scholar]
- 63.Georgantas R, Hildreth R. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc. Natl. Acad. Sci. 2007;104(8):2750–5. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc. Natl. Acad. Sci. 2007;104(17):7080–5. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiao C, Calado DP, Galler G, Thai T-H, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 66.Tan LP, Wang M, Robertus J-L, Schakel RN, Gibcus JH, Diepstra A, Harms G, Peh S-C, Reijmers RM, Pals ST, Kroesen B-J, Kluin PM, Poppema S, et al. miRNA profiling of B-cell subsets: specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab. Invest. 2009;89(6):708–16. doi: 10.1038/labinvest.2009.26. [DOI] [PubMed] [Google Scholar]
- 67.O'Connell RM, Chaudhuri A a, Rao DS, Gibson WSJ, Balazs AB, Baltimore D. MicroRNAs enriched in hematopoietic stem cells differentially regulate long-term hematopoietic output. Proc. Natl. Acad. Sci. U. S. A. 2010;107(32):14235–40. doi: 10.1073/pnas.1009798107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gibcus JH, Tan LP, Harms G, Schakel RN, Jong D, De Blokzijl T, Möller P, Poppema S, Kroesen B-J, van den Berg A. Hodgkin Lymphoma Cell Lines Are Characterized by a Specific miRNA Expression Profile. 2009;11(2):167–176. doi: 10.1593/neo.08980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thai T-H, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316(5824):604–8. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 70.Rodriguez A, Vigorito E, Clare S, Warren M V, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska E a, Vetrie D, Okkenhaug K, Enright AJ, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316(5824):608–11. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska E a, Rodriguez A, Bradley A, Smith KGC, Rada C, Enright AJ, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27(6):847–59. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Yébenes VG, Belver L, Pisano DG, González S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J. Exp. Med. 2008;205(10):2199–206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. microRNA-155 is a negative regulator of Activation Induced Cytidine deaminase. 2008;28(5):621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, Jaenisch R, Sharp P a, Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–86. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fernando TR, Rodriguez-Malave NI, Rao DS. MicroRNAs in B cell development and malignancy. J. Hematol. Oncol. 2012;5(1):7. doi: 10.1186/1756-8722-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.De Yébenes VG, Bartolomé-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunol. Rev. 2013;253(1):25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hanahan D, Weinberg RA, Francisco S. The Hallmarks of Cancer Review University of California at San Francisco. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 78.Hanahan D, Weinberg R a. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 79.Dimopoulos K, Gimsing P, Grønbæk K. Aberrant microRNA expression in multiple myeloma. Eur. J. Haematol. 2013;91(2):95–105. doi: 10.1111/ejh.12124. [DOI] [PubMed] [Google Scholar]
- 80.Pekarsky Y, Croce CM. Is miR-29 an Oncogene or Tumor Suppressor in CLL? 2010;1(3):224–227. doi: 10.18632/oncotarget.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. 2004;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baer C, Claus R, Frenzel LP, Zucknick M, Park YJ, Gu L, Weichenhan D, Fischer M, Pallasch CP, Herpel E, Rehli M, Byrd JC, Wendtner C-M, et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012;72(15):3775–85. doi: 10.1158/0008-5472.CAN-12-0803. [DOI] [PubMed] [Google Scholar]
- 84.Braconi C, Huang N, Patel T. MicroRNA-dependent regulation of DNA methyltransferase-1 and tumor suppressor gene expression by interleukin-6 in human malignant cholangiocytes. Hepatology. 2010;51(3):881–90. doi: 10.1002/hep.23381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duursma AM, Kedde M, Schrier M, Sage C le, Agami R. miR-148 targets human DNMT3b protein coding region. 2008;14:872–877. doi: 10.1261/rna.972008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. miR-449a targets HDAC-1 and induces growth arrest in prostate cancer. Oncogene. 2009;28(14):1714–24. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 87.Roccaro AM, Sacco A, Jia X, Azab AK, Maiso P, Ngo HT, Azab F, Runnels J, Quang P, Ghobrial IM. microRNA-dependent modulation of histone acetylation in Waldenstrom macroglobulinemia. Blood. 2010;116(9):1506–14. doi: 10.1182/blood-2010-01-265686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xiangjun H, Jing Y, Qi Z, Heng C, Yujun Z. Shortening of the 3′ untranslated region: an important mechanism leading to overexpression of HMGA2 in serous ovarian cancer. 2014;127(3):494–499. [PubMed] [Google Scholar]
- 89.Dabrowska MJ, Dybkaer K, Johnsen HE, Wang B, Wabl M, Pedersen FS. Loss of MicroRNA targets in the 3′ untranslated region as a mechanism of retroviral insertional activation of growth factor independence 1. J. Virol. 2009;83(16):8051–61. doi: 10.1128/JVI.00427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 91.Roccaro AM, Sacco A, Thompson B, Leleu X, Azab AK, Azab F, Runnels J, Jia X, Ngo HT, Melhem MR, Lin CP, Ribatti D, Rollins BJ, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113(26):6669–80. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hao M, Zhang L, An G, Meng H, Han Y, Xie Z, Xu Y, Li C, Yu Z, Chang H, Qiu L. Bone marrow stromal cells protect myeloma cells from bortezomib induced apoptosis by suppressing microRNA-15a expression. Leuk. Lymphoma. 2011;52(9):1787–94. doi: 10.3109/10428194.2011.576791. [DOI] [PubMed] [Google Scholar]
- 93.Zhou Y, Chen L, Barlogie B, Stephens O, Wu X, Williams DR, Cartron M-A, van Rhee F, Nair B, Waheed S, Pineda-Roman M, Alsayed Y, Anaissie E, et al. High-risk myeloma is associated with global elevation of miRNAs and overexpression of EIF2C2/AGO2. Proc. Natl. Acad. Sci. 2010;107(17):7904–9. doi: 10.1073/pnas.0908441107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Qian B, Katsaros D, Lu L, Preti M, Durando A, Arisio R, Mu L, Yu H. High miR-21 expression in breast cancer associated with poor disease-free survival in early stage disease and high TGF-beta1. Breast Cancer Res. Treat. 2009;117(1):131–40. doi: 10.1007/s10549-008-0219-7. [DOI] [PubMed] [Google Scholar]
- 95.Ma X, Yoshimoto K, Guan Y, Hata N, Mizoguchi M, Sagata N, Murata H, Kuga D, Amano T, Nakamizo A, Sasaki T. Associations between microRNA expression and mesenchymal marker gene expression in glioblastoma. Neuro. Oncol. 2012;14(9):1153–62. doi: 10.1093/neuonc/nos145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Liu Z-L, Wang H, Liu J, Wang Z-X. MicroRNA-21 (miR-21) expression promotes growth, metastasis, and chemo- or radioresistance in non-small cell lung cancer cells by targeting PTEN. Mol. Cell. Biochem. 2013;372(1-2):35–45. doi: 10.1007/s11010-012-1443-3. [DOI] [PubMed] [Google Scholar]
- 97.Hiyoshi Y, Kamohara H, Karashima R, Sato N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M, Baba H. MicroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin. Cancer Res. 2009;15(6):1915–22. doi: 10.1158/1078-0432.CCR-08-2545. [DOI] [PubMed] [Google Scholar]
- 98.Satzger I, Mattern A, Kuettler U, Weinspach D, Niebuhr M, Kapp A, Gutzmer R. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp. Dermatol. 2012;21(7):509–14. doi: 10.1111/j.1600-0625.2012.01510.x. [DOI] [PubMed] [Google Scholar]
- 99.Narducci MG, Arcelli D, Picchio MC, Lazzeri C, Pagani E, Sampogna F, Scala E, Fadda P, Cristofoletti C, Facchiano a, Frontani M, Monopoli a, Ferracin M, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sézary syndrome. Cell Death Dis. 2011;2(4):e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schramedei K, Mörbt N, Pfeifer G, Läuter J, Rosolowski M, Tomm JM, von Bergen M, Horn F, Brocke-Heidrich K. MicroRNA-21 targets tumor suppressor genes ANP32A and SMARCA4. Oncogene. 2011;30(26):2975–85. doi: 10.1038/onc.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balagué O, Gel B, Abrisqueta P, Lopez-Guillermo A, Artells R, Montserrat E, Monzo M. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111(5):2825–32. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- 102.Spizzo R, Nicoloso MS, Croce CM, Calin G. a. SnapShot: MicroRNAs in Cancer. Cell. 2009;137(3):586–586.e1. doi: 10.1016/j.cell.2009.04.040. [DOI] [PubMed] [Google Scholar]
- 103.Pichiorri F, Suh S-S, Ladetto M, Kuehl M, Palumbo T, Drandi D, Taccioli C, Zanesi N, Alder H, Hagan JP, Munker R, Volinia S, Boccadoro M, et al. MicroRNAs regulate critical genes associated with multiple myeloma pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 2008;105(35):12885–90. doi: 10.1073/pnas.0806202105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467(7311):86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 105.Leone E, Morelli E, Di Martino MT, Amodio N, Foresta U, Gullà A, Rossi M, Neri A, Giordano A, Munshi NC, Anderson KC, Tagliaferri P, Tassone P. Targeting miR-21 inhibits in vitro and in vivo multiple myeloma cell growth. Clin. Cancer Res. 2013;19(8):2096–106. doi: 10.1158/1078-0432.CCR-12-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eis PS, Tam W, Sun L, Chadburn A, Li Z, Gomez MF, Lund E, Dahlberg JE. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. 2005;102(10):3627–32. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Caramuta S, Lee L, Ozata DM, Akçakaya P, Georgii-Hemming P, Xie H, Amini R-M, Lawrie CH, Enblad G, Larsson C, Berglund M, Lui W-O. Role of microRNAs and microRNA machinery in the pathogenesis of diffuse large B-cell lymphoma. Blood Cancer J. 2013;3(January):e152. doi: 10.1038/bcj.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, Kroesen B-J, van den Berg A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005;207(2):243–9. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 109.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia high-grade lymphoma in Eμ-miR155 transgenic mice. Proc. Natl. Acad. Sci. 2006;103(18):7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol. 2010;42(8):1348–54. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ, Hammond SM. A microRNA polycistron as a potential human oncogene. Nature. 2005;435(7043):828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zanette DL, Rivadavia F, Molfetta G a, Barbuzano FG, Proto-Siqueira R, Silva W a, Jr, Falcão RP, Zago M a. miRNA expression profiles in chronic lymphocytic and acute lymphocytic leukemia. Braz. J. Med. Biol. Res. 2007;40(11):1435–40. doi: 10.1590/s0100-879x2007001100003. [DOI] [PubMed] [Google Scholar]
- 113.Li Y, Vecchiarelli-Federico LM, Li Y-J, Egan SE, Spaner D, Hough MR, Ben-David Y. The miR-17-92 cluster expands multipotent hematopoietic progenitors whereas imbalanced expression of its individual oncogenic miRNAs promotes leukemia in mice. Blood. 2012;119(19):4486–98. doi: 10.1182/blood-2011-09-378687. [DOI] [PubMed] [Google Scholar]
- 114.Schneider C, Setty M, Holmes AB, Maute RL, Leslie CS, Mussolin L, Rosolen A, Dalla-Favera R, Basso K. microRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc. Natl. Acad. Sci. 2014;111(22):8185–90. doi: 10.1073/pnas.1322466111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Felli N, Fontana L, Pelosi E, Botta R, Bonci D, Facchiano F, Liuzzi F, Lulli V, Morsilli O, Santoro S, Valtieri M, Calin GA, Liu C-G, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. 2005;102(50):18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Di Martino MT, Gullà A, Cantafio MEG, Lionetti M, Leone E, Amodio N, Guzzi PH, Foresta U, Conforti F, Cannataro M, Neri A, Giordano A, Tagliaferri P, et al. In Vitro and in Vivo Anti-tumor Activity of miR-221/222 Inhibitors in Multiple Myeloma. 2013;4(2):242–255. doi: 10.18632/oncotarget.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gottesman MM. Mechanisms of Cancer Drug Resistance. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- 118.Zheng T, Wang J, Chen X, Liu L. Role of microRNA in anticancer drug resistance. Int. J. Cancer. 2010;126(1):2–10. doi: 10.1002/ijc.24782. [DOI] [PubMed] [Google Scholar]
- 119.Allen KE, Weiss GJ. Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol. Cancer Ther. 2010;9(12):3126–36. doi: 10.1158/1535-7163.MCT-10-0397. [DOI] [PubMed] [Google Scholar]
- 120.Rizzo M, Mariani L, Cavallini S, Simili M, Rainaldi G. The over-expression of miR-34a fails to block DoHH2 lymphoma cell proliferation by reducing p53 via c-MYC down-regulation. Nucleic Acid Ther. 2012;22(4):283–8. doi: 10.1089/nat.2012.0343. [DOI] [PubMed] [Google Scholar]
- 121.Forte E, Salinas RE, Chang C, Zhou T, Linnstaedt SD, Gottwein E, Jacobs C, Jima D, Li Q-J, Dave SS, Luftig M a. The Epstein-Barr virus (EBV)-induced tumor suppressor microRNA MiR-34a is growth promoting in EBV-infected B cells. J. Virol. 2012;86(12):6889–98. doi: 10.1128/JVI.07056-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lodygin D, Tarasov V, Epanchintsev A, Berking C, Knyazeva T, Körner H, Diebold J, Hermeking H. Inactivation of miR-34a by aberrant CpG methylation in multiple types of cancer. Cell Cycle. 2008;7(16):2591–2600. doi: 10.4161/cc.7.16.6533. [DOI] [PubMed] [Google Scholar]
- 123.Craig VJ, Cogliatti SB, Imig J, Renner C, Neuenschwander S, Rehrauer H, Schlapbach R, Dirnhofer S, Tzankov A, Müller A. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117(23):6227–36. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Craig VJ, Tzankov a, Flori M, Schmid C a, Bader a G, Müller a. Systemic microRNA-34a delivery induces apoptosis and abrogates growth of diffuse large B-cell lymphoma in vivo. Leukemia. 2012;26(11):2421–4. doi: 10.1038/leu.2012.110. [DOI] [PubMed] [Google Scholar]
- 125.Di Martino MT, Leone E, Amodio N, Foresta U, Lionetti M, Pitari MR, Cantafio MEG, Gullà A, Conforti F, Morelli E, Tomaino V, Rossi M, Negrini M, et al. Synthetic miR-34a mimics as a novel therapeutic agent for multiple myeloma: in vitro and in vivo evidence. Clin. Cancer Res. 2012;18(22):6260–70. doi: 10.1158/1078-0432.CCR-12-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front. Genet. 2012;3(120):1–9. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sotillo E, Laver T, Mellert H, Schelter JM, Cleary M a, McMahon S, Thomas-Tikhonenko a. Myc overexpression brings out unexpected antiapoptotic effects of miR-34a. Oncogene. 2011;30(22):2587–94. doi: 10.1038/onc.2010.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rao E, Jiang C, Ji M, Huang X, Iqbal J, Lenz G, Wright G, Staudt LM, Zhao Y, McKeithan TW, Chan WC, Fu K. The miRNA-17~92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia. 2012;26(5):1064–72. doi: 10.1038/leu.2011.305. [DOI] [PubMed] [Google Scholar]
- 129.Todoerti K, Barbui V, Pedrini O, Lionetti M, Fossati G, Mascagni P, Rambaldi A, Neri A, Introna M, Lombardi L, Golay J. Pleiotropic anti-myeloma activity of ITF2357: inhibition of interleukin-6 receptor signaling and repression of miR-19a and miR-19b. Haematologica. 2010;95(2):260–9. doi: 10.3324/haematol.2009.012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bai H, Wei J, Deng C, Yang X, Wang C, Xu R. MicroRNA-21 regulates the sensitivity of diffuse large B-cell lymphoma cells to the CHOP chemotherapy regimen. Int. J. Hematol. 2013;97(2):223–31. doi: 10.1007/s12185-012-1256-x. [DOI] [PubMed] [Google Scholar]
- 131.Wang X, Li C, Ju S, Wang Y, Wang H, Zhong R. Myeloma cell adhesion to bone marrow stromal cells confers drug resistance by microRNA-21 up-regulation. Leuk. Lymphoma. 2011;52(10):1991–8. doi: 10.3109/10428194.2011.591004. [DOI] [PubMed] [Google Scholar]
- 132.Park S, Lee J, Ko YH, Han A, Jun HJ, Lee SC, Hwang IG, Park YH, Ahn JS, Jung CW, Kim K, Ahn YC, Kang WK, et al. The impact of Epstein-Barr virus status on clinical outcome in diffuse large B-cell lymphoma. Blood. 2007;110(3):972–8. doi: 10.1182/blood-2007-01-067769. [DOI] [PubMed] [Google Scholar]
- 133.Kim JH, Kim WS, Park C. Epstein-Barr virus latent membrane protein-1 protects B-cell lymphoma from rituximab-induced apoptosis through miR-155-mediated Akt activation and up-regulation of Mcl-1. Leuk. Lymphoma. 2012;53(8):1586–91. doi: 10.3109/10428194.2012.659736. [DOI] [PubMed] [Google Scholar]
- 134.Feng D-D, Zhang H, Zhang P, Zheng Y-S, Zhang X-J, Han B-W, Luo X-Q, Xu L, Zhou H, Qu L-H, Chen Y-Q. Down-regulated miR-331-5p and miR-27a are associated with chemotherapy resistance and relapse in leukaemia. J. Cell. Mol. Med. 2011;15(10):2164–75. doi: 10.1111/j.1582-4934.2010.01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shen H, Xu W, Luo W, Zhou L, Yong W, Chen F, Wu C, Chen Q, Han X. Upregulation of mdr1 gene is related to activation of the MAPK/ERK signal transduction pathway and YB-1 nuclear translocation in B-cell lymphoma. Exp. Hematol. 2011;39(5):558–69. doi: 10.1016/j.exphem.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 136.Yagi K, Yamamoto K, Umeda S, Abe S, Suzuki S, Onishi I, Kirimura S, Fukayama M, Arai A, Kitagawa M, Kurata M. Expression of multidrug resistance 1 gene in B-cell lymphomas: association with follicular dendritic cells. Histopathology. 2013;62(3):414–20. doi: 10.1111/his.12035. [DOI] [PubMed] [Google Scholar]
- 137.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 138.Stenvang J, Petri A, Lindow M, Obad S, Kauppinen S. Inhibition of microRNA function by antimiR oligonucleotides. Silence. 2012;3(1):1. doi: 10.1186/1758-907X-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Manfè V, Biskup E, Rosbjerg A, Kamstrup M, Skov AG, Lerche CM, Lauenborg BT, Odum N, Gniadecki R. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. PLoS One. 2012;7(1):e29541. doi: 10.1371/journal.pone.0029541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.He M, Gao L, Zhang S, Tao L, Wang J, Yang J, Zhu M. Prognostic significance of miR-34a and its target proteins of FOXP1, p53, and BCL2 in gastric MALT lymphoma and DLBCL. Gastric cancer. 2013 doi: 10.1007/s10120-013-0313-3. doi:10.1007/s10120-013-0313-3. [DOI] [PubMed] [Google Scholar]
- 141.Goswami RS, Atenafu EG, Xuan Y, Waldron L, Reis PP, Sun T, Datti A, Xu W, Kuruvilla J, Good DJ, Lai R, Church AJ, Lam WS, et al. MicroRNA Signature Obtained From the Comparison of Aggressive With Indolent Non-Hodgkin Lymphomas: Potential Prognostic Value in Mantle-Cell Lymphoma. J. Clin. Oncol. 2013;31(23):2903–11. doi: 10.1200/JCO.2012.45.3050. [DOI] [PubMed] [Google Scholar]
- 142.Wu P, Agnelli L, Walker B a, Todoerti K, Lionetti M, Johnson DC, Kaiser M, Mirabella F, Wardell C, Gregory WM, Davies FE, Brewer D, Neri A, et al. Improved risk stratification in myeloma using a microRNA-based classifier. Br. J. Haematol. 2013;162(3):348–59. doi: 10.1111/bjh.12394. [DOI] [PubMed] [Google Scholar]
- 143.Wu PY, Zhang XD, Zhu J, Gup XY, Wang JF. Low expression of microRNA-146b-5p and microRNA-320d predicts poor outcome of large B-cell lymphoma treated with cyclophosphamide, doxorubicin, vincristine, and prednisone. Hum. Pathol. 2014;45(8):1664–1673. doi: 10.1016/j.humpath.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 144.Visone R, Veronese A, Balatti V, Croce CM. MiR-181b: new perspective to evaluate disease progression in chronic lymphocytic leukemia. 2012;3(2):195–202. doi: 10.18632/oncotarget.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhu D-X, Zhu W, Fang C, Fan L, Zou Z-J, Wang Y-H, Liu P, Hong M, Miao K-R, Liu P, Xu W, Li J-Y. miR-181a/b significantly enhances drug sensitivity in chronic lymphocytic leukemia cells via targeting multiple anti-apoptosis genes. Carcinogenesis. 2012;33(7):1294–301. doi: 10.1093/carcin/bgs179. [DOI] [PubMed] [Google Scholar]
- 146.Lin S, Pan L, Guo S, Wu J, Jin L, Wang J-C, Wang S. Prognostic role of microRNA-181a/b in hematological malignancies: a meta-analysis. PLoS One. 2013;8(3):e59532. doi: 10.1371/journal.pone.0059532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Su R, Lin H-S, Zhang X-H, Yin X-L, Ning H-M, Liu B, Zhai P-F, Gong J-N, Shen C, Song L, Chen J, Wang F, Zhao H-L, et al. MiR-181 family: regulators of myeloid differentiation and acute myeloid leukemia as well as potential therapeutic targets. Oncogene. 2014;(July):1–14. doi: 10.1038/onc.2014.274. [DOI] [PubMed] [Google Scholar]
- 148.Lwin T, Lin J, Choi YS, Zhang X, Moscinski LC, Wright KL, Sotomayor EM, Dalton WS, Tao J. Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood. 2010;116(24):5228–36. doi: 10.1182/blood-2010-03-275925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Adams CM, Eischen CM. Inactivation of p53 Is Insufficient to Allow B Cells and B-Cell Lymphomas to Survive Without Dicer. Cancer Res. 2014;74(14):3923–34. doi: 10.1158/0008-5472.CAN-13-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sarasquete ME, Gutiérrez NC, Misiewicz-Krzeminska I, Paiva B, Chillón MC, Alcoceba M, García-Sanz R, Hernández JM, González M, San-Miguel JF. Upregulation of Dicer is more frequent in monoclonal gammopathies of undetermined significance than in multiple myeloma patients and is associated with longer survival in symptomatic myeloma patients. Haematologica. 2011;96(3):468–71. doi: 10.3324/haematol.2010.033845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Pellegrino L, Jacob J. Altered expression of the miRNA processing endoribonuclease Dicer has prognostic significance in human cancers. Expert Rev. Anticancer Ther. 2013;13(1):21–7. doi: 10.1586/era.12.150. [DOI] [PubMed] [Google Scholar]
- 152.Jones KL, Nourse JP, Keane C, Bhatnagar A, Gandhi M. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clin. cancer Res. an Off. J. Am. Assoc. Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-13-1024. doi:10.1158/1078-0432.CCR-13-1024. [DOI] [PubMed] [Google Scholar]
- 153.Sevcikova S, Kubiczkova L, Sedlarikova L, Slaby O, Hajek R. Serum miR-29a as a marker of multiple myeloma. Leuk. Lymphoma. 2013;54(1):189–91. doi: 10.3109/10428194.2012.704030. [DOI] [PubMed] [Google Scholar]
- 154.Song G, Gu L, Li J, Tang Z, Liu H, Chen B, Sun X, He B, Pan Y, Wang S, Cho WC. Serum microRNA expression profiling predict response to R-CHOP treatment in diffuse large B cell lymphoma patients. Ann. Hematol. 2014 doi: 10.1007/s00277-014-2111-3. doi:10.1007/s00277-014-2111-3. [DOI] [PubMed] [Google Scholar]
- 155.Williams Z, Ben-Dov IZ, Elias R, Mihailovic A, Brown M, Rosenwaks Z, Tuschl T. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proc. Natl. Acad. Sci. U. S. A. 2013;110(11):4255–60. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McDonald JS, Milosevic D, Reddi H V, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin. Chem. 2011;57(6):833–40. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 157.Chen W, Wang H, Chen H, Liu S, Lu H, Kong D, Huang X, Kong Q, Lu Z. Clinical significance and detection of microRNA-21 in serum of patients with diffuse large B-cell lymphoma in Chinese population. Eur. J. Haematol. 2014;92(5):407–12. doi: 10.1111/ejh.12263. [DOI] [PubMed] [Google Scholar]
- 158.Takei Y, Ohnishi N, Kisaka M, Mihara K. Determination of abnormally expressed microRNAs in bone marrow smears from patients with follicular lymphomas. Springerplus. 2014;3(288):1–9. doi: 10.1186/2193-1801-3-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bruchova H, Yoon D, Agarwal a M, Mendell J, Prchal JT. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35(11):1657–1667. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL, Rajewsky K. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 2008;9(4):405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Thapa DR, Li X, Jamieson BD, Martínez-Maza O. Overexpression of microRNAs from the miR-17-92 paralog clusters in AIDS-related non-Hodgkin's lymphomas. PLoS One. 2011;6(6):e20781. doi: 10.1371/journal.pone.0020781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo Y-M, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y, Shimizu N, Ichinohasama R, Sawada K. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114(15):3265–75. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 163.Wang F, Zhu Y, Guo L, Dong L, Liu H, Yin H, Zhang Z, Li Y, Liu C, Ma Y, Song W, He A, Wang Q, et al. A regulatory circuit comprising GATA1/2 switch and microRNA-27a/24 promotes erythropoiesis. Nucleic Acids Res. 2014;42(1):442–57. doi: 10.1093/nar/gkt848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rao DS, O'Connell RM, Chaudhuri A a, Garcia-Flores Y, Geiger TL, Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33(1):48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rodriguez-Ubreva J, Ciudad L, van Oevelen C, Parra M, Graf T, Ballestar E. C/EBPa-mediated activation of microRNAs 34a and 223 inhibits Lef1 expression to achieve efficient reprogramming into macrophages. Mol. Cell. Biol. 2014;34(6):1145–57. doi: 10.1128/MCB.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Ramkissoon SH, Mainwaring L a, Ogasawara Y, Keyvanfar K, McCoy JP, Sloand EM, Kajigaya S, Young NS. Hematopoietic-specific microRNA expression in human cells. Leuk. Res. 2006;30(5):643–7. doi: 10.1016/j.leukres.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 167.Yu S-C, Chen S-U, Lu W, Liu T-Y, Lin C-W. Expression of CD19 and lack of miR-223 distinguish extramedullary plasmacytoma from multiple myeloma. Histopathology. 2011;58(6):896–905. doi: 10.1111/j.1365-2559.2011.03793.x. [DOI] [PubMed] [Google Scholar]
- 168.Guo L, Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS One. 2010;5(6):e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]