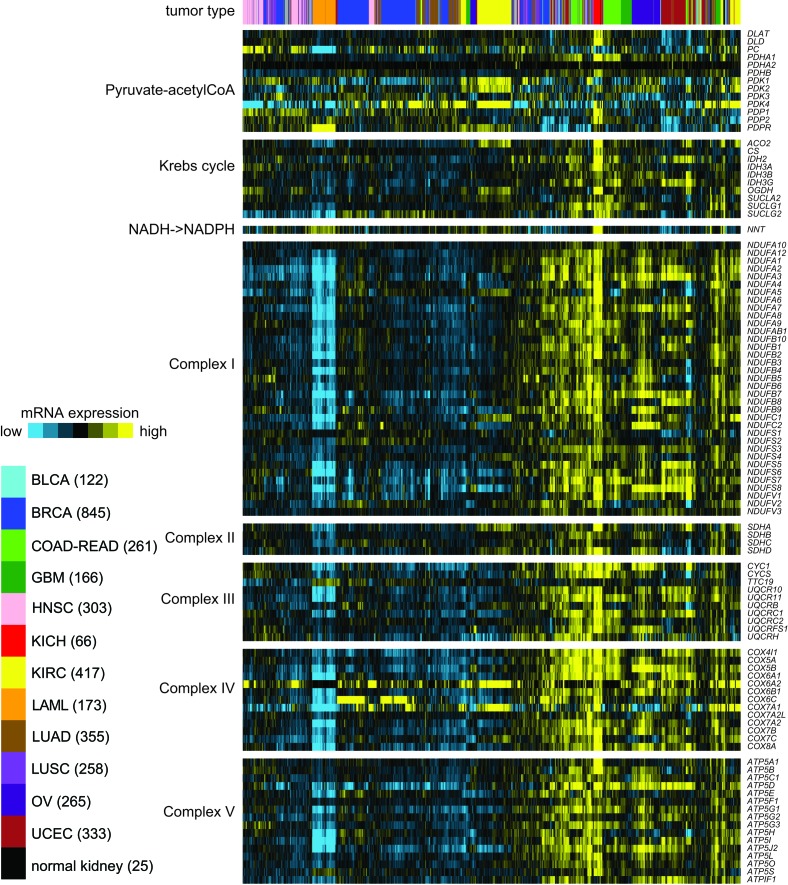
Figure 1. In a panel of human cancers of various tissues of origin (from TCGA, n = 3,564 samples), differential expression (relative to sample median) of genes related to Krebs cycle and Electron Transport Chain (ETC).
RNA-seq profiles are from the recent TCGA pan-cancer multiplatform subtyping analysis, with the addition of sample profiles of ChRCC (KICH) and normal kidney from TCGA ChRCC study [1]. On the basis of the expression patterns of the selected genes, samples were clustered by unsupervised hierarchical method [50]. KICH, chromophobe renal cell carcinoma (ChRCC); BLCA, bladder cancer; BRCA, breast cancer; COAD, colon cancer; GBM, glioblastoma; HNSC, head and neck squamous cell carcinoma; KIRC, clear cell renal cell carcinoma (ccRCC); LAML, acute myeloid leukemia; LUAD, lung adenocarcinoma; LUSC, lung squamous; OV, serous ovarian cancer; READ, rectal cancer; UCEC, endometrial cancer.