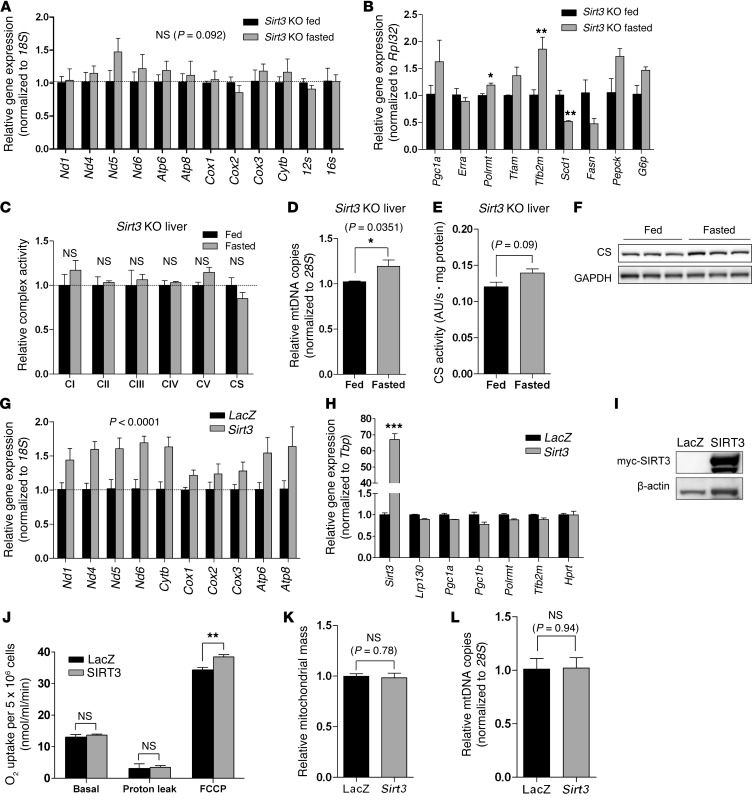
Figure 4. In liver, SIRT3 is necessary and sufficient for fasting-mediated induction of mitochondrial encoded transcripts and OXPHOS.
(A) Hepatic gene expression of mitochondrially encoded transcripts in 24-hour fasted Sirt3 KO mice (129S background) (n = 3). (B) Expression of genes that regulate mitochondrial biogenesis, mitochondrial transcription, lipogenesis, and gluconeogenesis in Sirt3 KO liver (n = 3). (C) Complex activity from liver of 24-hour fasted or fed Sirt3 KO mice (n = 3). (D) Genetic assessment of mitochondrial content using mtDNA content (n = 3). (E) Biochemical assessment of mitochondrial content using citrate synthase activity in whole liver homogenate (n = 3). (F) Assessment of mitochondrial content by immunoblotting citrate synthase protein in whole liver homogenate (n = 3). (G and H) Expression of (G) mitochondrially encoded transcripts and (H) genes that regulate mitochondrial biogenesis and mitochondrial transcription in H2.35 hepatoma cells stably expressing LacZ or SIRT3 (n = 3). (I) Immunoblot showing expression of ectopically expressed myc-tagged SIRT3 protein. (J) Using a Clark-type oxygen electrode, cellular respiration — basal, proton leak, and maximal respiration (FCCP) — was assessed in H2.35 hepatoma cells stably expressing LacZ or SIRT3. (n = 4). See Supplemental Figure 6 for representative oxygen consumption curves. (K) Fluorescent assessment of mitochondrial content using MitoTracker Green FM (n = 3). (L) Genetic assessment of mitochondrial content using mtDNA content (n = 3). Data are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, 2-way ANOVA, with (A and G) or without (B and H) Bonferroni post-test, or 2-tailed unpaired Student’s t test (C–E and J–L).