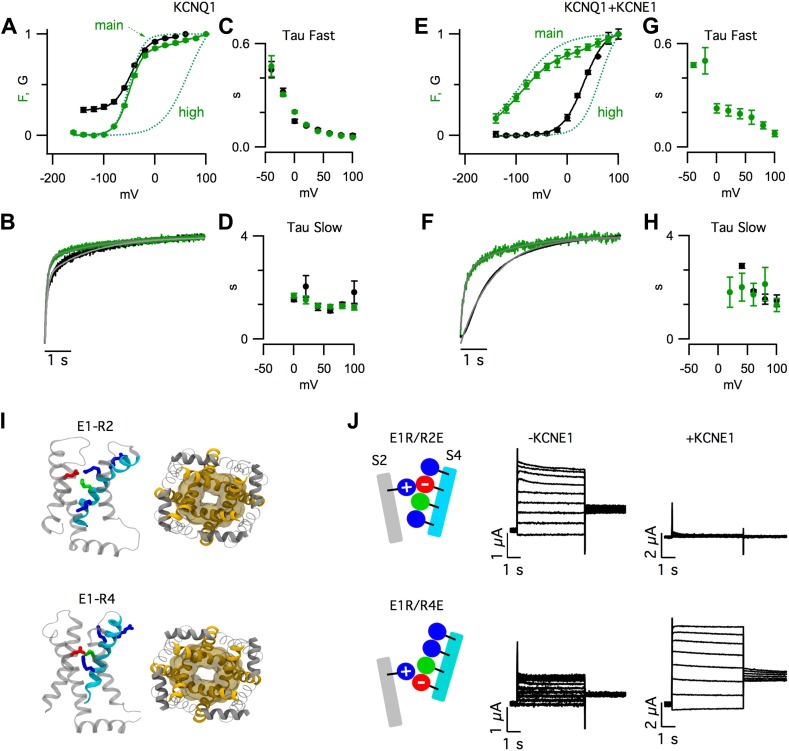
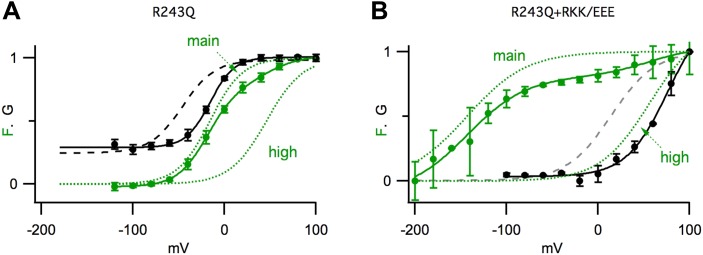
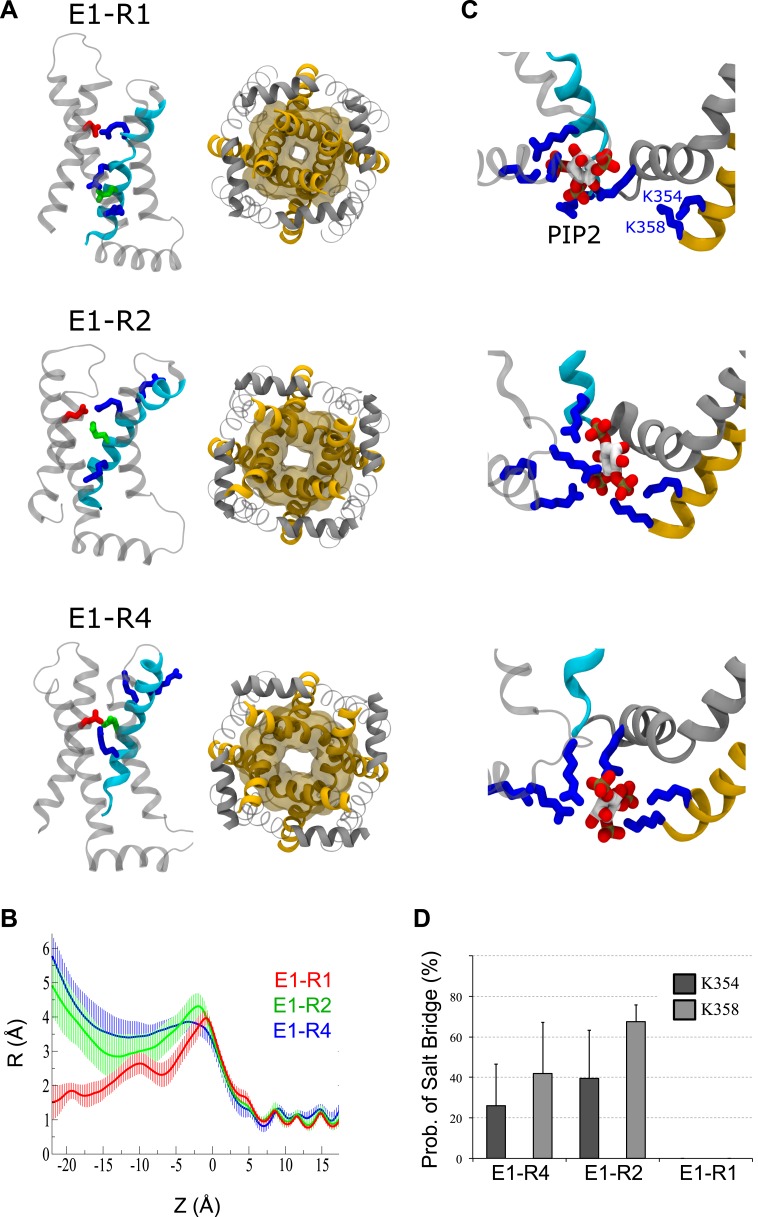
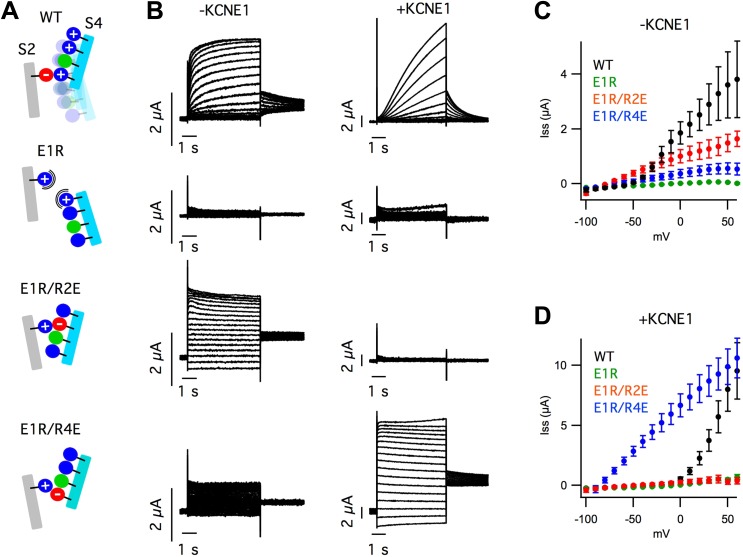
(
A) Snapshots of one VSD (left, side view) or the pore domain
(right, bottom view) following 100 ns of MD simulations. In the resting,
intermediate or activated VSD, E160 (E1, red) forms a salt bridge with R228
(R1), R231 (R2) or R237 (R4), respectively. (
B) Averaged (over
several trajectories) pore radius vs position along the axis normal to the
membrane (Z). (
C) Snapshots of the PIP
2
intrasubunit-binding site in the three states. In the resting/closed state,
PIP
2 interacts with positive residues of S4 (cyan). When the
VSD is intermediate or activated, PIP
2 shifts closer to S6
(yellow) and anchors its positive residues (K354 and K358). (
D)
Probability of salt bridges formation between positive residues of S6 (K354
and K358) and PIP
2. The lipid interacts with S6 only when the VSD
is intermediate or activated, not when it is resting. Error bars represent
SD. K354 and K358 interactions are not statistically different for the E1-R2
and E1-R4 states.
Figure 1—figure
supplement 3B represents the averaged pore radius profiles along
the axis normal to the membrane (Z). In the activated/open and intermediate
states, the minimal radiuses of the pore at this level are 3.5 ± 0.4
Å and 2.8 ± 0.6 Å respectively. For comparison, in the Kv1.2
open state, the corresponding radius (pdb 3LUT [
Chen et al., 2010]) is 4.2 Å, in the Kv1.2/2.1
paddle chimera open state (pdb 2 R9R [
Long
et al., 2007]) it is 4.2 Å also, and in the NavMS open state
(pdb 3ZJZ [
Loussouarn et al., 2003])
it is 2.3 Å. Therefore, the minimal pore radius at the intercellular
gate level in the models of the Kv7.1 activated and intermediate states
corresponds to the open pore. In the resting/closed state, this radius
decreases to 1.5 ± 0.5 Å. This is similar to the closed states of
KcsA (pdb 1K4C [
Zhou et al.,
2001]), NavAB (pdb 4EKW [
Payandeh
et al., 2012]) and NavAP (pdb 4DXW [
Zhang et al., 2012]), where these values are 1.1, 1.2
and 0.9 Å respectively. The activated/open, intermediate and
resting/closed states of Kv7.1 differ by their properties as evidenced from
the reported experimental data. Taking advantage of our simulations, we
attempted to investigate whether the interactions between PIP
2
and positive residues of the Kv7.1 intrasubunit binding site are different.
Indeed PIP
2 interacts preferably with the VSD (S4) when the
channel is resting/closed or with the pore (S6) when the channel is
activated/open (
Kasimova et al.,
2014) (
Figure 1—figure
supplement 3C, top and bottom panels). In the intermediate state,
the lipid forms salt bridges with both S4 (R243) and S6 (K354 and K358)
simultaneously (
Figure 1—figure
supplement 3C, middle panel). Its equilibrium position is also
between these in the activated/open and resting/closed states.
Interestingly, the probability of interaction between PIP
2 and S6
(K354 and K358) is rather high (
Figure
1—figure supplement 3D). The average values are slightly
higher for the intermediate than for the activated/open states: 40 and 26%
for K354, 68 and 42% for K358 respectively. However, this difference is
statistically insignificant due to the estimated error bars.