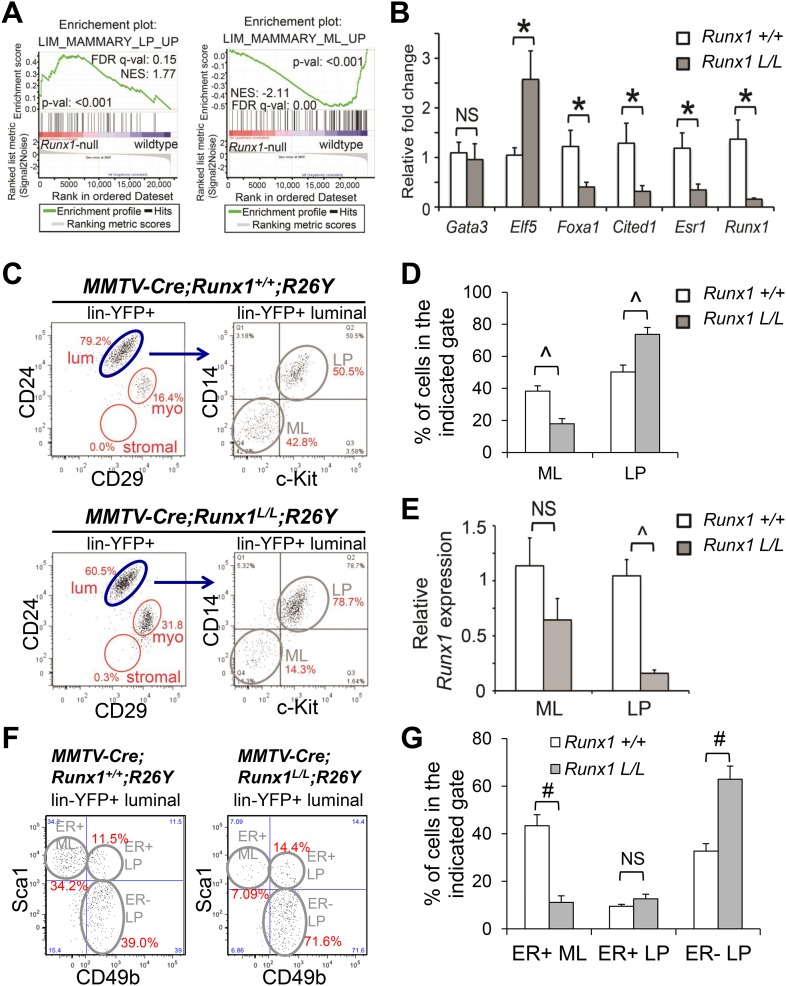
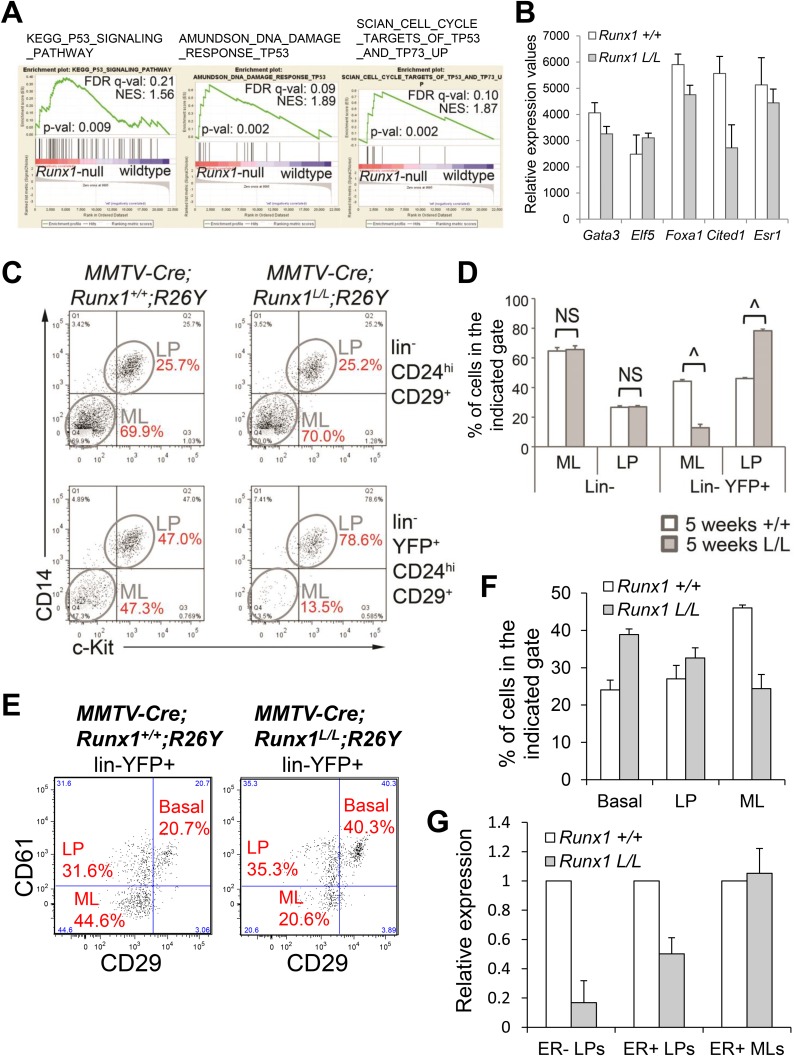
Figure 4. Runx1 disruption leads to a profound reduction in ER+ MLs.
(A) GSEA enrichment plots showing correlation of the expression profiles of Runx1-null or WT luminal MECs with previously published conserved human and mouse signatures of LPs (left) or MLs (right) (Lim et al., 2010). (B) qRT-PCR validation of TF/co-factor genes known to play roles in luminal lineage specification and maintenance. RNA was isolated from YFP+ Runx1-null and WT primary luminal MECs. (C) FACS plots of expression of CD14 and c-Kit, two LP markers (Asselin-Labat et al., 2011), in the gated YFP+ luminal MECs (Lin−CD24+CD29lo) of adult MMTV-Cre;Runx1L/L;R26Y virgin females and MMTV-Cre;Runx1+/+;R26Y control females. Note the CD14−c-Kit− mature luminal (ML) subpopulation was largely lacking in the lower right plot. (D) Quantification of the percentages of the ML and LP subpopulations as indicated in C, showing significant reduction in the ML subpopulation in MMTV-Cre;Runx1L/L;R26Y females (n = 13) compared to that in MMTV-Cre;Runx1+/+;R26Y control females (n = 10). (E) qRT-PCR analysis showing significantly reduced Runx1 expression in the LP subpopulation but not in the ML subpopulation in MMTV-Cre;Runx1L/L;R26Y females. (F) FACS plots of expression of CD49b, a LP marker, and Sca1, an ER+ ML marker (Shehata et al., 2012) in the gated YFP+ luminal MEC population. Note the CD49b−Sca1+ ER+ ML subpopulation was dramatically reduced, whereas the CD49b+Sca1− ER− LP subpopulation was increased in MMTV-Cre;Runx1L/L;R26Y females. (G) Quantification of the percentages of the ER+ ML, ER+ LP, and ER− LP subpopulations as indicated in F, showing significant reduction in the ER+ ML subpopulation in MMTV-Cre;Runx1L/L;R26Y females (n = 4) compared to those in MMTV-Cre;Runx1+/+;R26Y control females (n = 4). p values: *: p ≤ 0.05; #: p ≤ 0.005; ^: p ≤ 0.0005; NS = not significant; error bars represent mean ± S.E.M.
DOI: http://dx.doi.org/10.7554/eLife.03881.008