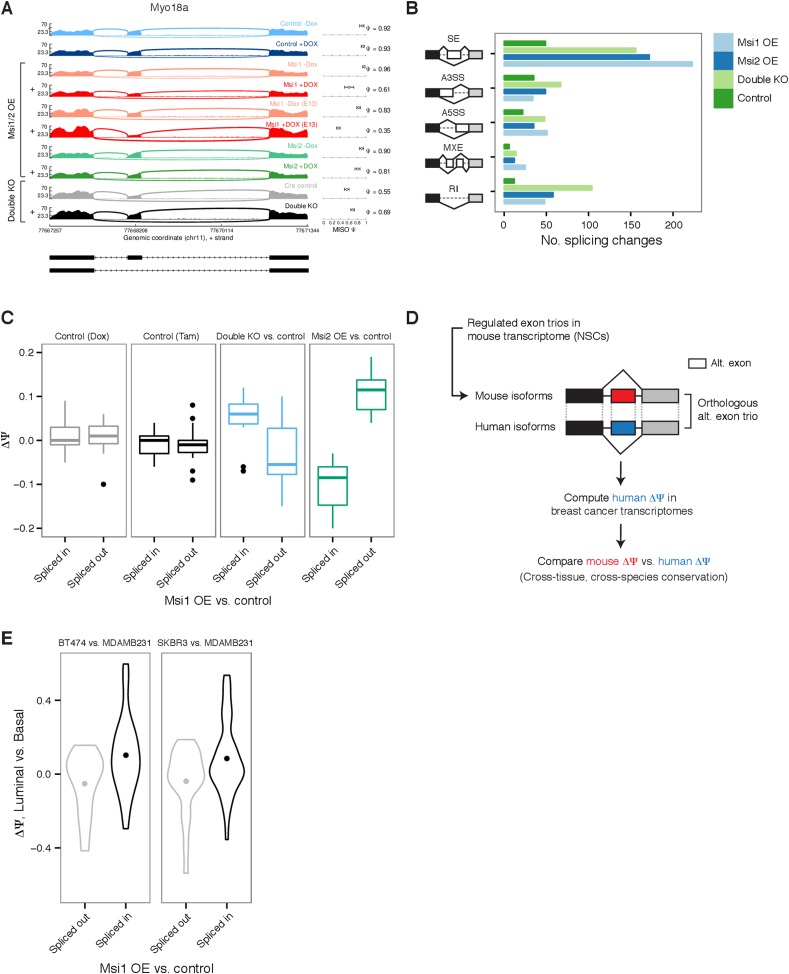
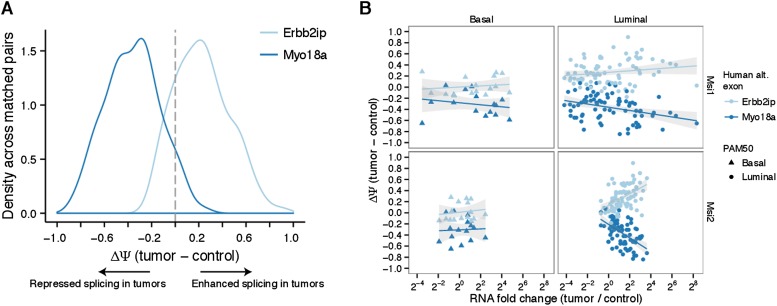
Figure 5. Global impact of Msi proteins on alternative splicing.
(A) Sashimi plot for Myo18a alternative exon 38 with Percent Spliced In (Ψ) estimates by MISO (values with 95% confidence intervals, right panel.) Exon splicing is repressed by Msi1 overexpression and slightly increased in knockout Msi1/2 cells. ‘+’ indicates samples treated with Dox/Tam for overexpression/knockout cells, respectively. E12.5 neural stem cells were used for all samples except Msi1 overexpression for which an additional E13.5 NSC time point was sequenced. (B) Number of differential events (MISO Bayes factor ≥10, ΔΨ ≥ 0.12) in each alternative RNA processing category (SE—skipped exons, A5SS—alternative 5′ splice site, A3SS—alternative 3′ splice site, MXE—mutually exclusive exons, RI—retained introns) for Msi1 overexpression (‘Msi1 OE’), Msi2 overexpression (‘Msi2 OE’), double knockouts (‘Double KO’), and a Dox control pair (‘Control’). (C) Comparison of ΔΨ in Msi1 overexpression vs control binned by direction (‘Spliced in’ or ‘Spliced out’, x-axis) to ΔΨ in Msi2 overexpression cells and in double knockout cells (along with respective Tam and Dox controls, y-axis). (D) Computational strategy for identifying human orthologs of alternative exon trios regulated in mouse neural stem cells. Orthologous exon trios were identified by synteny using multiple genome alignments. (E) Comparison of ΔΨ mouse alternative exons by Msi1 (comparing overexpression to control, x-axis) and ΔΨ of their orthologous exon trios in human (comparing luminal and basal cell lines, y-axis). Two pairs of luminal and basal cells compared: BT474 vs MDAMB231 and SKBR3 vs MDAMB231. ΔΨ value distributions summarized by violin plots with a dot indicating the mean ΔΨ value.