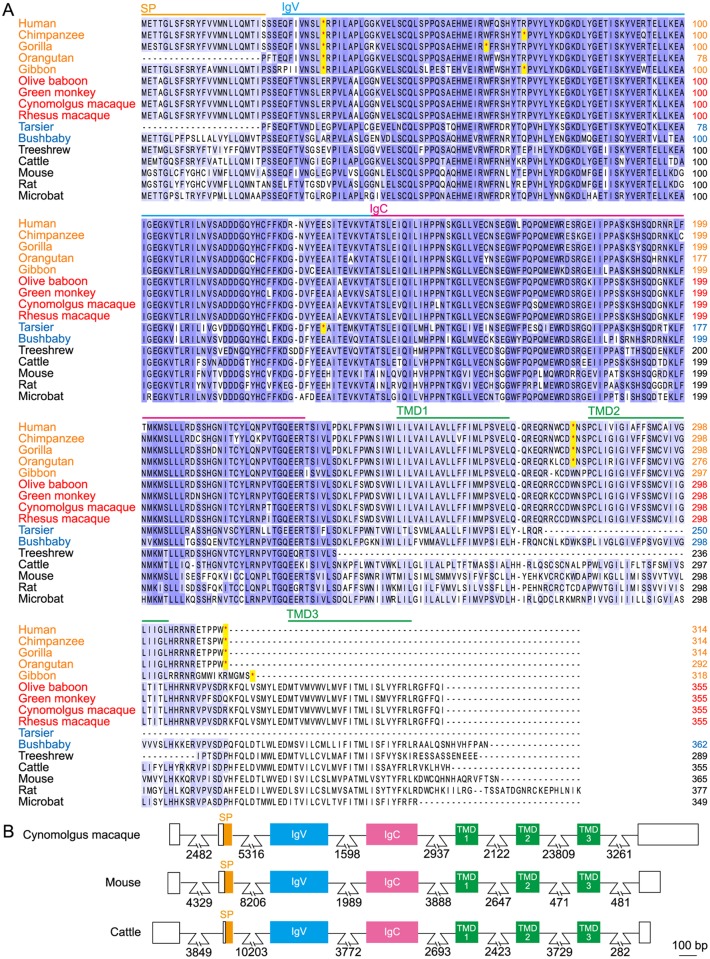
Fig 1. Amino acid sequence alignment of SKINT1 and SKINT1L molecules.
A. Deduced SKINT1 and SKINT1L protein sequences of indicated species were aligned using the Clustal Omega program. Strictly and highly conserved residues are indicated in dark blue and light blue, respectively. Hominoid sequences contain multiple premature stop codons (indicated by red asterisks). The location of predicted domains is indicated on top of the sequences. Species names indicated in orange, red and blue represent hominoids, OWMs and prosimians, respectively. SP stands for signal peptides. Accession numbers are as follows: cattle, XP_005204826; microbat, XP_006107913.1; mouse, NP_001096132; rat, NP_001129388; and treeshrew, XP_006147332.1. Accession numbers and relevant information for primate sequences are given in S1 Table. B. The exon-intron organization of cynomolgus macaque SKINT1L was compared to that of mouse Skint1 and bovine SKINT1L. Exon-intron boundaries were predicted based on the consensus splice junction sequences and similarity of the deduced amino acid sequences to the mouse SKINT1 protein sequence. Numbers indicate the length of introns in base pairs. Open boxes indicate 5'- and 3'-untranslated regions. bp, base pairs.