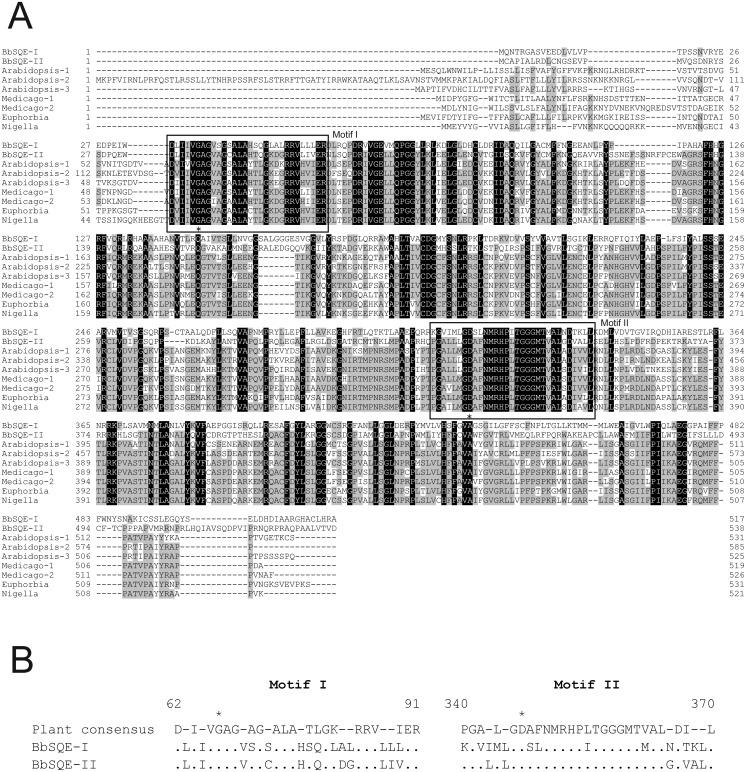
Fig 2. Amino acid sequences of BbSQEs-I and -II aligned with those from the other organisms.
A. The amino acid sequences of land plant SQEs that complemented Saccharomyces cerevisiae erg1 and BbSQEs were aligned using CLUSTAL W (ver.1.83) multiple sequence alignment tool (www.ebi.ac.uk/Tools/msa/clustalw2/help/faq.html, accessed Nov. 14, 2014) and adjusted manually. Amino acid residues that are 100% identical in the alignment are highlighted in black and those which are more than 50% identical are highlighted in grey. Boxes show conserved domains, hyphens denote the gaps in aligned sequences and asterisks indicate amino acid residues whose point mutations could result in the loss of complementation of erg1 [26]. B. Sequences of motifs I and II in BbSQEs-I and -II aligned with plant consensus sequence. The top sequence shows the consensus among seven plant sequences from panel A. Numbers denote amino acid residue positions in AtSQE1. Hyphens indicate varieties in plant sequences. Asterisks are the same as in panel A. BbSQE sequences that are identical to the plant consensus are shown with dots.