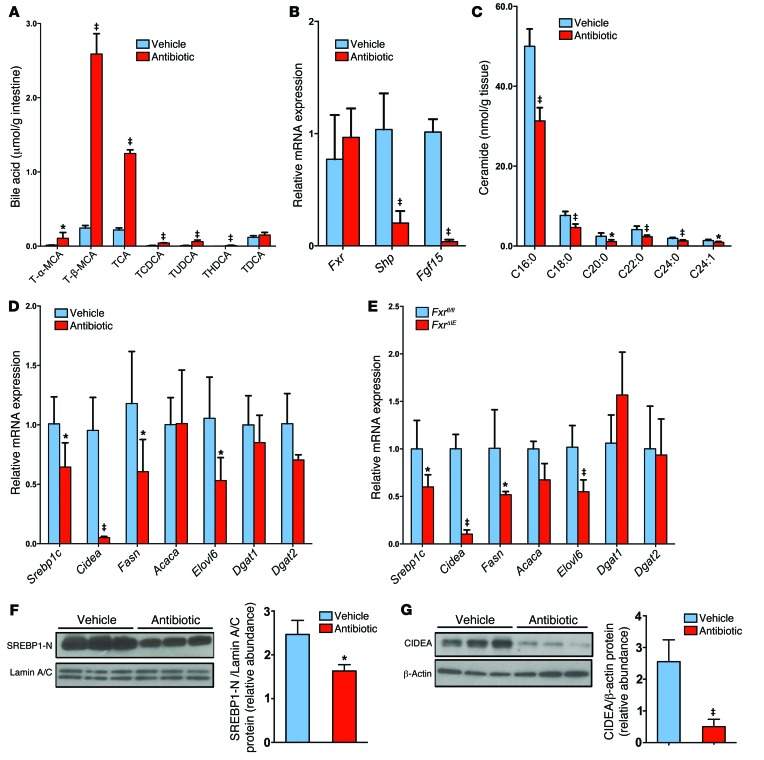
Figure 6. Intestinal FXR signaling influences the ceramide/SREBP1C/CIDEA pathway.
(A) Ileal bile acid profiles after 3 days of antibiotic treatment in HFD-fed mice. n = 5 mice per group. (B) Fxr, Shp, and Fgf15 mRNA levels in the ileum after 3 days of antibiotic treatment in HFD-fed mice. n = 5 mice per group. (C) Ileal ceramide profiles after 3 days of antibiotic treatment in HFD-fed mice. n = 5 mice per group. (D) mRNA levels of fatty acid synthesis– and triglyceride synthesis–related enzymes in the liver of HFD-fed mice after 7 weeks of antibiotic treatment. n = 5 mice per group. (B–D) Expression was normalized to 18S RNA. (A–D) Data are presented as the mean ± SD. *P < 0.05 and ‡P < 0.01 (2-tailed Student’s t test) compared with vehicle-treated mice. (E) mRNA levels for encoding enzymes involved in fatty acid and triglyceride synthesis in the livers of Fxrfl/fl and FxrΔIE mice fed a HFD for 14 weeks. n = 5 mice per group. Expression was normalized to 18S RNA. Data are presented as the mean ± SD. *P < 0.05 and ‡P < 0.01 (2-tailed Student’s t test) compared with Fxrfl/fl mice. (F) Western blot analysis of liver nuclear SREBP1-N expression after 7 weeks of antibiotic treatment in HFD-fed mice and quantitation of SREBP1-N expression. n = 3 mice per group. (G) Western blot analysis of liver CIDEA expression after 7 weeks of antibiotic treatment in HFD-fed mice and quantitation of SREBP1-N expression. n = 3 mice per group. (F and G) Data are presented as the mean ± SD. *P < 0.05 and ‡P < 0.01 (2-tailed Student’s t test) compared with vehicle-treated mice.