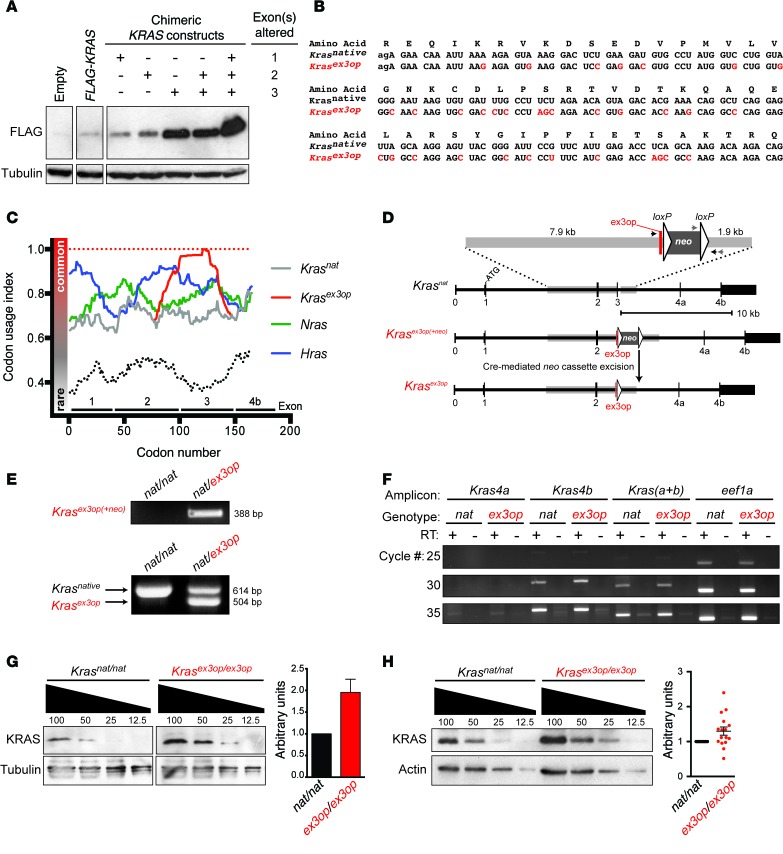
Figure 1. Generation and characterization of the Krasex3op allele.
(A) Immunoblot reveals changing rare codons to common codons in exon 3 increases ectopic KRAS expression (1 of 2 experiments using cells independently transduced with expression vectors). (B) Position of synonymous mutations (red) in Krasex3op aligned to Krasnat within exon 3. Lowercase letters denote nucleotides from exon 2. (C) Sliding window of codon usage index demonstrating the relative codon usage of the mouse Ras genes including the engineered enrichment of common codons in exon 3 of Krasex3op (red) relative to Krasnat (gray). Theoretical Kras transcripts encoded by all common (red dotted line) or all rare (black dotted line) are plotted for reference. (D) Targeting strategy to change codon usage in exon 3 of Kras. Arrows indicate PCR primers used for genotyping. (E) Representative genotyping from mice with Krasnat and/or Krasex3op alleles used in all genotyping experiments. (F) Semiquantitative RT-PCR analysis revealed similar levels of Kras splice forms 4a and 4b in MEFs isolated from Krasex3op/ex3op versus those from Krasnat/nat mice (1 of 2 experiments using independently derived MEFs). (G) Representative example and quantification (mean ± SEM) of immunoblots revealed higher levels of KRAS protein in lysates of MEFs isolated from Krasex3op/ex3op than levels detected in Krasnat/nat mice (1 of 2 experiments using independently-derived MEFs). (H) Representative example and quantification (mean ± SEM) of immunoblots revealed higher levels of KRAS protein in total lung lysates isolated from Krasex3op/ex3op mice than those detected in Krasnat/nat mice. Comparisons were made using lysates from a total of 16 Krasex3op/ex3op versus 16 Krasnat/nat mice.