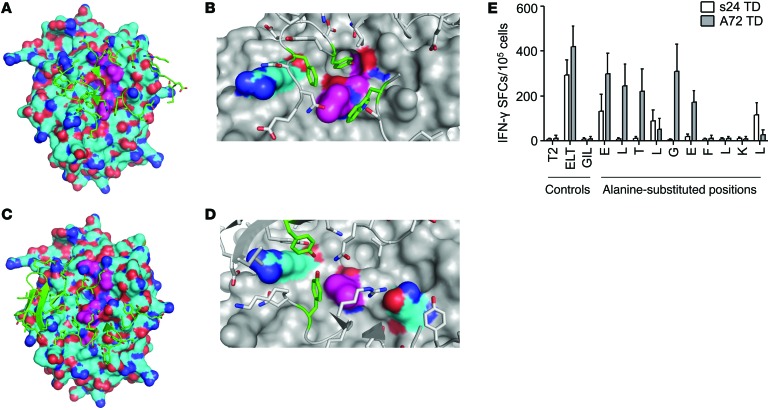
Figure 8. Different molecular recognition patterns of autologous versus allogeneic repertoire–derived survivin-specific TCRs.
(A–D) Structural modeling reveals a highly epitope-specific binding interface of the novel autologous survivin-specific TCR s24. (A and C) HLA-A*0201 (cyan) and survivin ELT peptide (magenta) are represented at the surface; TCR regions directly in contact with the HLA-peptide complex are represented in green. (B and D) Residues with the most energetically favorable contacts across the binding interface are in green (TCR), cyan (HLA) or magenta (ELT peptide). (B) Trp 289, Tyr 352, and Phe 356 from the s24-TCR, Leu 4, Gly 5, and Phe 7 from the survivin peptide, and Arg 65 from the HLA. (D) Trp 244, Tyr 290 from the A72-TCR, Leu 4 from the survivin peptide, and Arg 65 and Gln 155 from the HLA. (E) Alanine substitution analysis testing s24 TD (white bars) or A72 TD (gray bars) T cells for recognition of peptide-pulsed T2 cells by IFN-γ ELISpot assay. Data represent the mean ± SD (n = 4 donors).