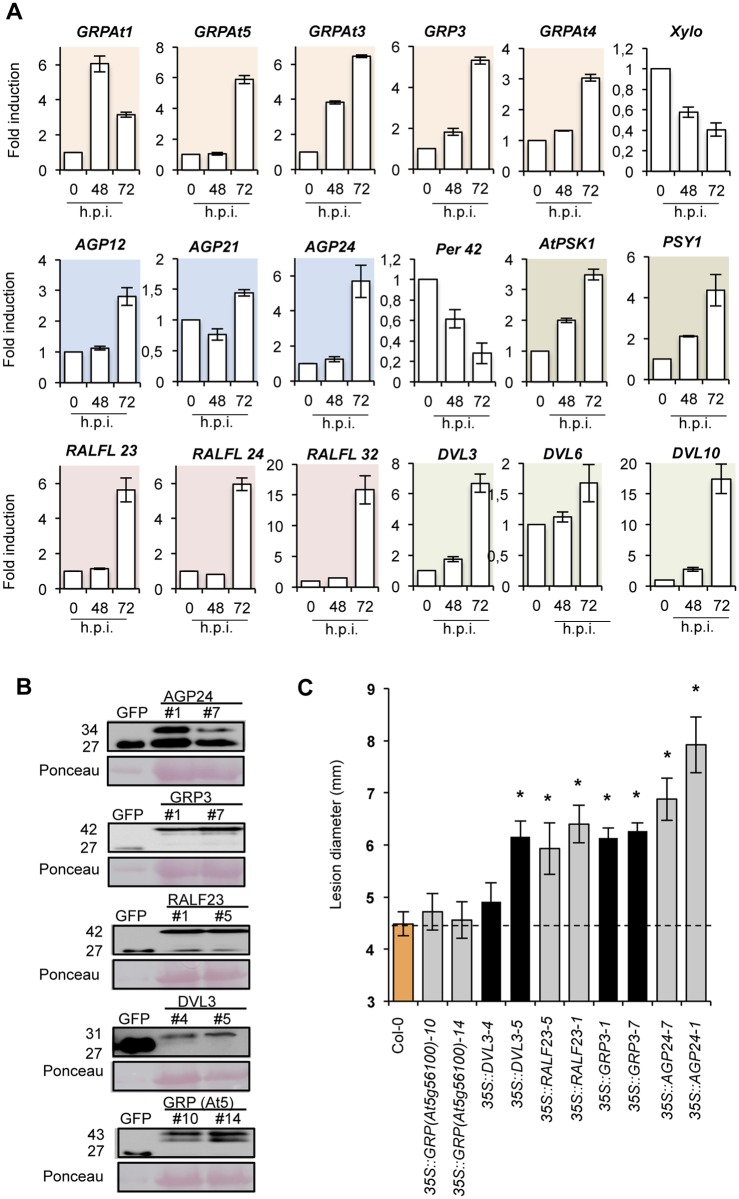
Fig 4. Functional implication of different signaling peptides in Col-0 disease response to P. cucumerina infection.
(A) Expression of genes encoding distinct members of GRP, AGP, PSK1, PSY, RALF, and DVL signaling peptides in Col-0 plants at early P. cucumerina infection stages. Relative expression was assayed over a 72-h time course by quantitative RT-PCR on total RNA from leaves following inoculation with a drop of P. cucumerina spore suspension. Expression of the Per42 peroxidase and the xyloglucan:xyloglucosyl transferase Xylo genes was concurrently assayed as internal controls and used for comparison. Data represent means ± SD (n = 3 biological replicates). Expression was normalized to the constitutive ACT2 gene, then to expression at each time point in mocked Col-0 plants. (B) Western blots with anti-GFP antibodies of crude protein extracts derived from T3 homozygous Col-0 plants expressing either a 35S::GFP construct or AGP24-GFP, GRP3-GFP, RALF23-GFP, DVL3-GFP and GRP (At5g56100)-GFP constructs. Two independent lines for each gene construct were used and the accumulation of the encoded fusion protein compared to that of free GFP. Ponceau staining of the nitrocellulose filter confirmed equal protein loading. (C) Resistance response to P. cucumerina of Col-0 and two independent homozygous lines expressing each of the signaling peptides proteins fused to GFP shown in panel B. Disease was evaluated 11 d.p.i by determining the average lesion diameter on three leaves per plants and from 15 plants per genotype. Data points represent average lesion size ± SE of measurements. An ANOVA was conducted to assess significant differences in disease symptoms with a 0.05 level of significance. Error bars represent standard deviation (SD) (n = 12). Asterisks indicate statistical significant differences.