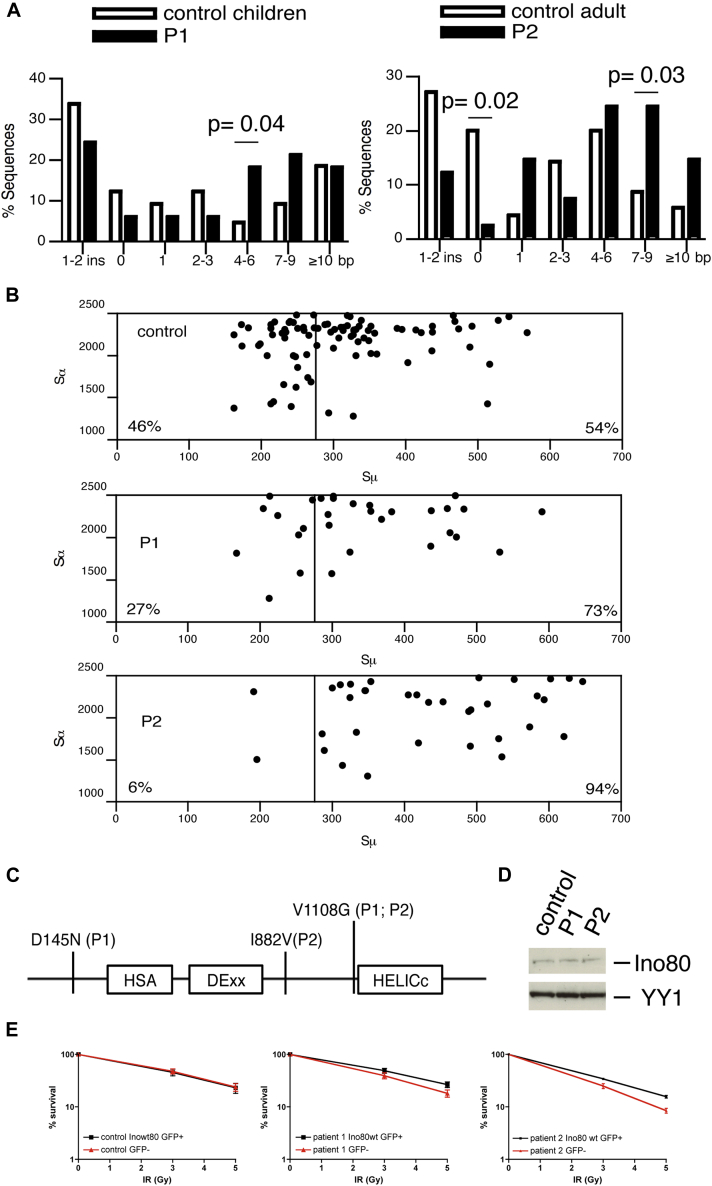
Fig 1.
A, Abnormal switch junction repair in 2 patients carrying INO80 gene variations. Analysis of Sμ–Sα recombination junctions. White bars indicate control sequences (65 and 70 for children and adult controls, respectively, recently published37). Black bars indicate patient sequences (33 and 41 for P1 and P2, respectively). B, Scatterplot analysis of Sμ and Sα breakpoints. Vertical line at position 275 indicates the start of the Sμ region with highest degree of homology with Sα1 and Sα2. C, INO80 protein structure. D, Immunoblot analysis of INO80 and YY1. Radiosensitivities of lentivirally infected patients and control fibroblast cell lines (E) co-expressing wt INO80 and GFP; P value from paired Student t test of percentage survival at 5 Gy for patients' INO80wt GFP+ cells versus patients' GFP− cells for P1: .02 and for P2: .04.