Abstract
Based on the observation of reduced stature in relatives of patients with acromesomelic dysplasia, Maroteaux type (AMDM), caused by homozygous or compound heterozygous mutations in natriuretic peptide receptor-B gene (NPR2), it has been suggested that heterozygous mutations in this gene could be responsible for the growth impairment observed in some cases of idiopathic short stature (ISS). We enrolled 192 unrelated patients with short stature and 192 controls of normal height and identified seven heterozygous NPR2 missense or splice site mutations all in the short stature patients, including one de novo splice site variant. Three of the six inherited variants segregated with short stature in the family. Nine additional rare nonsynonymous NPR2 variants were found in three additional cohorts. Functional studies identified eight loss-of-function mutations in short individuals and one gain-of-function mutation in tall individuals. With these data we were able to rigorously verify that NPR2 functional haploinsufficiency contributes to short stature. We estimate a prevalence of NPR2 haploinsufficiency of between 0 and 1/26 in people with idiopathic short stature. We suggest that NPR2 gain of function may be a more common cause of tall stature than previously recognized.
Keywords: Natriuretic peptide receptor-B, NPR2, short stature, ISS
Introduction
C-type natriuretic peptide (CNP, gene natriuretic peptide precursor C, NPPC; MIM# 123830) is a small, secreted peptide and a member of the natriuretic peptide family. CNP binds to a homodimeric transmembrane receptor named natriuretic peptide receptor-B (NPR-B, gene NPR2; MIM# 108961), which functions as a guanylyl cyclase to generate cGMP in chondrocytes, female reproductive organs, and endothelial cells (Schulz, 2005; Potter et al., 2006). In the growth plate chondrocytes, binding of CNP to NPR-B stimulates chondrocyte differentiation and hypertrophy as well as increases matrix synthesis. This occurs in part through NPR-B signaling inhibiting MAP kinase signaling by FGFR3 (Mericq et al., 2000; Yasoda et al., 2004; Krejci et al., 2005).
Several lines of evidence indicate that CNP/NPR-B signaling is an important regulator of skeletal growth. CNP-overexpressing mice exhibit excessive growth (Yasoda et al., 2004), while defects of the Nppc (Chusho et al., 2001) or Npr2 (Tamura et al., 2004) gene lead to impairment of skeletal development. In humans, biallelic loss-of-function mutations in NPR2 cause acromesomelic dysplasia, Maroteaux type (AMDM; MIM# 602875). This autosomal recessive skeletal dysplasia is characterized by dwarfism and short limbs (Bartels et al., 2004). On the other hand, overproduction of CNP due to a chromosomal translocation has been reported in association with a skeletal dysplasia characterized by tall stature (Bocciardi et al., 2007; Moncla et al., 2007). In addition, gain-of-function mutations of NPR2 have been identified in several families and are associated with an overgrowth disorder with only mild skeletal features (Miura et al., 2012; Hannema et al., 2013; Robinson et al., 2013; Miura et al., 2014).
Interestingly, in the first report of biallelic NPR2 mutations causing AMDM, parents of patients with AMDM (obligate heterozygotes) were noted to be shorter than expected for their population of origin (Bartels et al., 2004), although these individuals came from a wide range of geographic and ethnic backgrounds, which complicated the interpretation of these findings. Another study that evaluated a single family with an AMDM proband showed that the heterozygous carriers had a mean height 1.4 SD lower than their non-carrier family members (Olney et al., 2006). In this single-family study, the proband’s parents share a common ancestor, so it is possible that the heterozygous carriers share some other mutation causing short stature, and the level of evidence of a heterozygous effect was limited by the size of the family. Based on these two studies, it is presumed that heterozygous NPR2 loss-of-function mutations can mildly impair long bone growth. It has further been hypothesized that one person in 30 with idiopathic short stature (ISS) will carry a NPR2 mutation (Bartels et al., 2004; Olney et al., 2006).
Recent studies searched for heterozygous NPR2 mutations in cohorts with ISS (Table 1). One study of 47 independent Brazilian patients identified heterozygous NPR2 mutations in 6% of patients (Vasques et al., 2013). Another study on 101 unrelated Japanese patients with short stature identified heterozygous NPR2 mutations in 2% of patients (Amano et al., 2014). While providing observational data consistent with the hypothesis that a monoallelic NPR2 mutation could cause short stature, these studies did not include controls without short stature, and were based on a relatively small number of patients. Hence, the hypothesis that NPR2 haploinsufficiency leads to short stature has not been rigorously verified. Analyses of larger cohorts, including controls, are needed to more clearly define the role of heterozygous NPR2 defects in patients with ISS.
Table 1.
Summary of recent studies searching for heterozygous NPR2 mutations in cohorts with ISS
| Cohort | # of functional NPR2 variants |
# of ISS individuals |
Fraction of ISS individuals with functional NPR2 variants |
Reference |
|---|---|---|---|---|
| Brazilian patients with ISS | 3 | 47 | 6% | (Vasques et al. 2013) |
| Japanese patients with short stature | 2 | 101 | 2% | (Amano et al. 2014) |
Materials and Methods
Subjects
Short stature patient cohort and Framingham Heart Study controls
192 patients with short stature (height more than 2 SD below the mean for age and sex) (Kuczmarski et al., 2002) but without defined etiology, were recruited from the endocrinology and genetics clinics at Boston Children's Hospital. 104 subjects fit strict criteria for ISS with no evidence of growth hormone deficiency, significant medical comorbidity, developmental delay, or suspected syndromic cause for their short stature. Of these 104 patients, height data was available for both parents in 99 cases. 22 of these 99 subjects (22.2%) had at least one parent with a height below −2 SD and were defined as having familial ISS. In addition, 192 control subjects were chosen from the middle of the Framingham Heart Study (FHS) height distribution (height z scores between −0.7 and +0.7). More detailed description of the cohort is reported in Wang, et al. (Wang et al., 2013)
Second short stature patient cohort
216 patients were taken from the Genetics and Neuroendocrinology of Short Stature International Study (GeNeSIS), sponsored by Eli Lilly and Company and various pediatric endocrinology clinics in Jacksonville, FL and Santiago, Chili. Patients were diagnosed by the treating provider with idiopathic short stature and had height < −2 SD below the mean for age and sex.
FINRISK height extreme cohort
272 subjects were chosen from the extremes (<1st percentile or >99th percentile) of the FINRISK surveys (FINRISK 1992, FINRISK 1997, FINRISK 2002, FINRISK 2007) height distribution. The FINRISK cohorts comprise the respondents of representative, cross-sectional population surveys that are carried out every 5 years since 1972, to assess the risk factors of chronic diseases and health behavior in the working age population, in five large study areas of Finland (Vartiainen et al., 2010).
Estonian Biobank height extreme cohort
1000 subjects were selected from the extremes (<1.25th percentile or >98.75th percentile) of the Estonian Biobank height distribution. The Estonian Biobank cohort is a volunteer-based sample of the Estonian resident adult population (age ≥18 years). The age, sex and geographical distribution closely reflect those of the Estonian adult population and encompass close to 5% of the entire adult population of Estonia (Leitsalu et al., 2014).
Pooled sequencing protocol
For the first short stature cohort and the FINRISK and Estonian cohorts, DNA samples from multiple subjects were pooled for DNA sequencing using previously described methods available at the Broad Institute (Rivas et al., 2011). To identify variants present only in a single individual (hereafter referred to as singleton variants), we applied a simple overlapping pooling design as described in Wang, et al. (Wang et al., 2013).
Sequencing was performed on the Illumina HiSeq platform. Variant calling was performed using a likelihood-based calling strategy we developed as described previously (Wang et al., 2013). Variants were annotated for functional effect using SnpEff 2.0.5 (http://snpeff.sourceforge.net/). The reference sequence used for numbering nucleotide change and amino acid change was NM_003995.3 and ENSP00000341083. Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence, with the initiation codon as codon 1.Variant allele frequency data was obtained from two publicly available databases: 1) Integrated variant call set of 1000 Genomes samples (Abecasis et al., 2010) (May 2013 release); 2) National Heart, Lung, and Blood Institute Exome Variant Server (National Heart, Lung). Maximal allele frequency from the two sources was used. Novel variants are those not observed in either of these databases. Confirmation of variants found in NPR2 through pooled sequencing was done via Sanger sequencing or Sequenom MassArray genotyping. Each variant was sequenced in the proband and in all related family members for whom DNA samples were provided. Sanger sequencing of NPR2 was performed in the second short stature cohort. For details of all sequencing protocols, see the Supp. Methods. The variants reported have been submitted to http://www.lovd.nl/NPR2.
In silico prediction of mutation effects
To identify the potential effects of sequence variants identified in NPR2 on protein function or structure, the wild-type and variant sequences were assessed using PolyPhen-2 (software version 2.2.2, http://genetics.bwh.harvard.edu/pph2) (Ramensky et al., 2002). The options used were: Classifier model [HumDiv], Genome aseembly [GRCh37/hg19], Transcripts [CCDS], Annotations [All].
Assaying wild-type and mutant NPR2 activity
Missense mutations in NPR2 were generated by site-directed mutagenesis kit (Agilent Technologies) using the wild-type rat NPR2 expression construct pRK5-NPR-B. Activity in HEK 293T cells was measured by at least one of two assays, one on whole cell lysate and another enzyme based assay done on membrane fractions of transfected cells, as described elsewhere (Yoder et al., 2008; Robinson and Potter, 2011). For details of the assay, see the Supp. Methods. Variants showing loss of activity in vitro in either assay were characterized as loss-of-function. Variants showing normal in vitro activity in the whole cell lysate assay were tested in the more sensitive enzyme based assay. Variants showing clear increases in activity in both assays were characterized as gain-of-function.
Results
Discovery of NPR2 variants in patients with short stature and controls using pooled sequencing
We selected 192 patients with short stature (more than 2 SD below the mean for age and sex) and 192 controls of matching ancestry from the middle of the Framingham Heart Study height distribution (height z scores between −0.7 and +0.7). Characteristics of the subjects were described previously (Wang et al., 2013); 104 subjects fit strict criteria for ISS with no evidence of growth hormone deficiency, significant medical comorbidity, developmental delay, or suspected syndromic cause for their short stature. Pooled targeted sequencing of the exons of 1077 candidate genes was performed (Wang et al., 2013). We detected 11 variants in NPR2. Of these, two were synonymous SNPs found in multiple patients and controls, and two were synonymous variants found only in patients. We focused on the seven potential loss-of-function variants, which included one splice site and six missense variants, all found in patients only (Table 2). These seven variants were all validated via traditional Sanger sequencing and confirmed to be heterozygous in each individual carrier.
Table 2.
NPR2 potentially nonsynonymous variants in short stature patients
| Cohort | Subject | Sex | Variant | Minor Allele Frequency |
PolyPhen2 Prediction |
Nadir Height SDS |
Current Height SDS |
GH Therapy |
IGF-1 Level (ng/ml) |
|---|---|---|---|---|---|---|---|---|---|
| Short stature patient cohort | Patient 1 | M | c.1352-1G>A | Novel | N/A | −3.14 | −2.15 | Yes | 53 @ 7 yr (44–211) |
| Patient 2 | M | c.142G>T (p.A48S) (brachyolmia with known TRPV4 variant) | Novel | 1.00 | −4.15 | −3.91 | No | NA | |
| Patient 3 | F | c.1167G>T (p.E389D) | Novel | 0.00 | −3.84 | −3.14 | Yes | 53 @ 6 yr (47–217) | |
| Patient 4 | F | c.1481T>G (p.I494S) | Novel | 0.997 | −3.57 | −1.79 | Yes | 115@ 6 yr (47–217) | |
| Patient 5 | M | c.1645G>A (p.A549T) | Novel | 0.988 | −3.88 | −2.70 | Yes | 34 @ 4 yr (25–157) | |
| Patient 6 | M | rs62637657: c.491C>G (p.A164G) | 0.02% | 0.065 | −2.53 | −2.16 | IGF-1 therapy | 72 @ 9 yr (61–252) | |
| Patient 7 | F | rs114147262: c.2359C>T (p.R787W) | 0.16% | 0.998 | −2.92 | −2.41 | No | 61 @ 7 yr (55–238) | |
| Second short stature cohort | Patient | NA | c.316G>T (p.A106S) | Novel | 0.189 | N/A | N/A | N/A | N/A |
| 3 Patients | NA | rs180950551: c.1802G>C (p.R601P) | 0.02% | 1.00 | N/A | N/A | N/A | N/A | |
| Patient | NA | c.2710A>T (p.K904*) | Novel | N/A | N/A | N/A | N/A | N/A | |
| Patient | NA | rs114115939: c.1517G>A (p.R506H) | 0.06% | 0.959 | N/A | N/A | N/A | N/A |
Abbreviations: F, female; M, male; GH, growth hormone; IGF-1, insulin-like growth factor 1; yr, years old; N/A, not available.
Normal age and gender specific ranges for IGF-1 are provided in parentheses.
GenBank reference sequence used for NPR2 is NM_003995.3.
Family analyses and clinical phenotypes
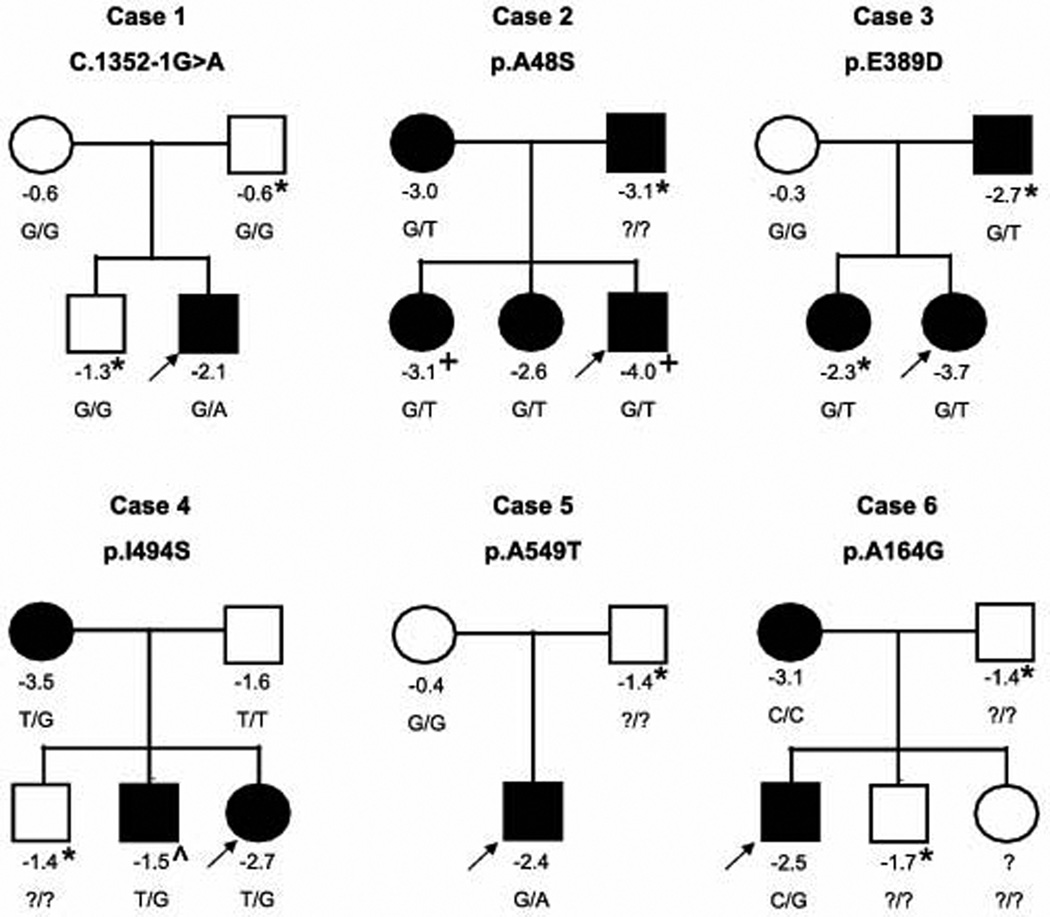
The splice site mutation is c.1352-1G>A in a highly conserved base pair in the acceptor splice site at the 5’ end of exon 7, carried in a single patient (Patient 1, Table 2). Sanger sequencing of the proband, his parents, and a brother confirmed that the variant is found in the patient but not in his mother, father or brother. These results were confirmed by a second round of sequencing, and paternity was confirmed by SNP genotyping. There was no family history of short stature (Figure 1), consistent with the hypothesis that the de novo variant in NPR2 is contributing to short stature in this patient.
Figure 1. Segregation of identified NPR2 nonsynonymous variants in affected families.
Numbers below the individuals denote the height z scores. + indicates family members who also carry the TRPV4 mutation causal of brachyolmia. ^ indicates that this family member was treated with growth hormone therapy prior to this height measurement and had a nadir height of −2.8 SDS. * indicates that the height was estimated by a family member. All other values were measured. Individuals with short stature are indicated as black circles (females) or squares (males). The arrow points to the affected proband in each family.
We also detected six missense variants in NPR2; of these, four were private variants not found in the public databases (Patients 2–5, Table 2). Sequencing of relatives demonstrated that three of these novel variants segregated with the short stature phenotype (Patients 2–4, Figure 1). The mother of Patient 5, who did not have short stature, did not carry the NPR2 variant, but the father’s DNA was not available to further confirm the segregation. Of note, one of the three segregating variants (Patient 2) was found in a male patient who was also found to carry a known TRPV4 mutation (c.1858G>A (p.V620I)) previously reported as causal for brachyolmia type 3 (Rock et al., 2008). The patient carries a clinical diagnosis of brachyolmia and features consistent with this disease, including platyspondyly of the cervical spine. We previously reported the presence of this likely pathogenic variant in this patient (Wang et al., 2013). The NPR2 variant was present in the patient’s mother, who does not carry the TRPV4 variant, and in two sisters, the elder of which also carries the TRPV4 variant. The mother and both sisters had short stature with height SDS scores below −2.5, and the sister carrying both variants had a height SDS score of −3.1. Notably the patient’s father is deceased but also was reported to have had short stature (−3.1 SDS) and presumably carried the TRPV4 variant. Thus, this patient and one of his sisters inherited the NPR2 variant from his mother and the TRPV4 variant from his father, presumably resulting in severe short stature with skeletal dysplasia.
We also detected two rare missense variants present in the public databases (Patients 6–7). Neither of these variants has been reported to be pathogenic, and both are reported to be relatively rare (minor allele frequency 0.02% and 0.16%). The mother of Patient 6, who also had short stature, did not carry the NPR2 variant, but DNA was not available from the sibling or father for further segregation analysis. Patient 7 was adopted from China, so the family history of short stature was unknown and testing for segregation of the variant was not possible.
Discovery of NPR2 variants in one additional clinical short stature cohort
To further refine the estimate of the fraction of ISS individuals with functional NPR2 variants, we screened for rare nonsynonymous NPR2 variants in a second short stature cohort (n=216) from the Lilly GeNeSIS Study and a number of pediatric endocrinology clinics. Patients were diagnosed by the treating provider with ISS and had height more than 2 SD below the mean for age. The exons and intron/exon boundaries of the NPR2 gene were sequenced using Sanger sequencing.
Four nonsynonymous variants in the coding region were identified, one nonsense and three missense variants (Table 2). One of the missense variants (rs180950551: c.1802G>C (p.R601P)) was seen in three separate patients. An in silico analysis suggested that one variant (c.316G>T (p.A106S)) is benign and the other two (rs180950551: c.1802G>C (p.R601P) and rs114115939: c.1517G>A (p.R506H)) are probably damaging.
Screening for NPR2 mutations in population-based cohorts of individuals from the extremes of the height distribution
We then went on to screen for rare nonsynonymous NPR2 variants in two additional cohorts. The first cohort of individuals (n=272) was selected from the extremes (<1st percentile or >99th percentile) of height distribution from four FINRISK surveys (~33,000 samples in total). Pooled targeted sequencing of the exons of 1077 candidate genes was performed. We detected three variants in NPR2. Of these variants, one was a common synonymous SNP (observed in the short stature patients and FHS controls as well), which we did not pursue further. The other two were missense variants, which we validated by genotyping and confirmed to be heterozygous in tall extremes only (Table 3). Neither variant was found in the public databases. An in silico analysis suggested that one mutation is benign (c.739A>G (p.N247D)) and the other is probably damaging (c.1685G>A (p.R562Q)).
Table 3.
NPR2 potentially nonsynonymous variants in FINRISK and Estonian Biobank height extreme samples
| Variant | Observation | Minor Allele Frequency |
PolyPhen2 Prediction |
|---|---|---|---|
| c.739A>G (p.N247D) | FINRISK tall extreme (also observed in Estonian tall extreme and short extreme) | Novel | 0.002 |
| c.1685G>A (p.R562Q) | FINRISK tall extreme | Novel | 0.995 |
| c.2449G>A (p.E817K) | Estonian short extreme | Novel | 0.609 |
| c.2338G>A (p.G780R) | Estonian short extreme | Novel | 0.013 |
| c.1982C>A (p.T661K) | Estonian short extreme | Novel | 0.999 |
GenBank reference sequence used for NPR2 is NM_003995.3.
The second cohort of individuals (n=1,000) was taken from the extremes (<1.25th percentile or >98.75th percentile) of height distribution from the Estonian Biobank (~52,000 samples in total). Pooled targeted sequencing of the exons of four candidate genes was performed. As before, we did not attempt to validate or further evaluate the potential synonymous or intronic variants. One missense variant (c.739A>G (p.N247D)), previously identified in one FINRISK tall extreme sample, was observed in both tall extreme and short extreme samples. The remaining three missense variants (Table 3) were each found in single short individuals. These allelic variants were not found in the public databases. An in silico analysis suggested that the three missense mutations are possibly damaging (c.2449G>A (p.E817K)), benign (c.2338G>A (p.G780R)), and probably damaging (c.1982C>A (p.T661K)), respectively.
Functional characterization of NPR2 variants
Overall, across four cohorts, we observed and validated 16 NPR2 nonsynonymous variants (14 missense, 1 nonsense and 1 splice site), 14 in short stature samples, 1 in a tall extreme sample, 1 which was present in both short and tall extreme samples, and none in normal height controls (Table 4). To further examine the functional consequences of the identified NPR2 nonysnonymous variants, we assessed the CNP-dependent cGMP-producing capacities of the 14 missense variants by transfecting HEK 293T cells, which do not normally express NPR2, using at least one of two methods. Relative guanylyl cyclase activities (±SD) of these variants compared with wild type are listed in Table 5. The whole cell lysate method was less sensitive at detecting partial loss of function mutations. The de novo splice variant (c.1352-1G>A) and the nonsense variant (c.2710A>T (p.K904*)) were not tested and are presumed to be loss of function variants.
Table 4.
Observation of NPR2 nonsynonymous variants in four cohorts
| Cohort | Observation of nonsynonymous variants |
Note |
|---|---|---|
| Short stature patients and FHS controls | 7 variants in 192 short stature patients 0 variant in 192 FHS controls |
Patient 1: de novo mutation Patients 2–4: all heterozygous relatives had short stature (height SDS < −2) Patient 5: variant not present in average height mother and father’s DNA unavailable Patient 6: variant not present in short mother and father’s DNA unavailable Patient 7: no family data available |
| Second short stature cohort | 4 variants in 216 short stature patients | No control samples |
| Extremes of FINRISK height distribution | 0 variant in 136 short extremes 2 variants in 136 tall extremes |
No family data available |
| Extremes of Estonian Biobank height distribution | 3 variants in 500 short extremes 0 variant in 500 tall extremes |
No family data available |
Table 5.
Functional characterization of NPR2 nonsynonymous variants in three cohorts
| Variant | Observation | Minor Allele Frequency |
Segregation Within Family |
Relative Guanylyl Cyclase Activities % (±SD) – Enzyme Assay with 1uM CNP |
Relative Guanylyl Cyclase Activities % (±SD) – Whole Cell Lysate |
|---|---|---|---|---|---|
| c.1352-1G>A | Patient 1 | Novel | de novo | N/A | N/A |
| c.142G>T (p.A48S) | Patient 2 | Novel | Yes | 36.6 (14.9)* | 93.5 (15.4) |
| c.1167G>T (p.E389D) | Patient 3 | Novel | Yes | 57.0 (12.0)* | 108.5 (24.6) |
| c.1481T>G (p.I494S) | Patient 4 | Novel | Yes | <6* | 54.3 (8.6)* |
| c.1645G>A (p.A549T) | Patient 5 | Novel | Unknown | 68.7 (26.9) | 111.2 (9.6) |
| rs62637657: c.491C>G (p.A164G) | Patient 6 | 0.02% | No | 25.9 (10.3)* | 130.6 (15.5)* |
| rs114147262: c.2359C>T (p.R787W) | Patient 7 | 0.16% | N/A | 62.8 (36.6) | 90.9 (14.0) |
| c.316G>T (p.A106S) | Patient | Novel | N/A | 83.9 (32.5) | N/A |
| rs180950551: c.1802G>C (p.R601P) | Patient | 0.02% | N/A | 64.5 (12.9) | N/A |
| c.2710A>T (p.K904*) | Patient | Novel | N/A | N/A | N/A |
| rs114115939: c.1517G>A (p.R506H) | Patient | 0.06% | N/A | <6* | N/A |
| c.739A>G (p.N247D) | FINRISK tall extreme (also observed in Estonian tall extreme and short extreme) | Novel | N/A | 69.1 (37.3) | 246.9 (66.5)* |
| c.1685G>A (p.R562Q) | FINRISK tall extreme | Novel | N/A | 145.7 (31.6)* | 711.8 (238.2)* |
| c.2449G>A (p.E817K) | Estonian short extreme | Novel | N/A | N/A | 31.1 (0.3)* |
| c.2338G>A (p.G780R) | Estonian short extreme | Novel | N/A | N/A | 98.3 (11.9) |
| c.1982C>A (p.T661K) | Estonian short extreme | Novel | N/A | N/A | 65.7 (19.1)* |
p <0.05.
GenBank reference sequence used for NPR2 is NM_003995.3.
To characterize the variants as likely nonpathogenic, variants of unknown significance, and likely pathogenic, we considered both the functional results and the pattern of segregation in the families, where available. For the six missense variants identified in the first short stature patient cohort, three of them segregate with short stature (Patient 2–4). For the variants in Patient 2–4, we observed decreased functional activity in at least one of the assays, strongly suggesting that these are pathogenic variants. The variant from patient 4 also showed decreased expression by Western blot (Supp. Figure S1). For the variant in Patient 5, the segregation is unknown and the functional data are negative. The variant in Patient 7, a known rare population variant, also had unchanged functional activity. We conclude that these two variants are unlikely to be pathogenic. The variant in Patient 6, which showed a slight increase in activity in the whole cell lysate assay and decreased activity in the enzyme based assay, was characterized as variant of unknown significance. Interestingly, three of the pathogenic variants were found amongst the 22 cases of familial ISS thus providing a diagnostic yield of 13.6%. However, only a single pathogenic variant, the de novo variant, was found amongst the 77 cases of non-familial ISS leading to a much lower diagnostic yield of 1.3%.
One of three missense variants in the second clinical short stature cohort (rs114115939: c.1517G>A (p.R506H)) decreased NPR-B activity likely secondary to decreased expression of the mutated receptor as seen on Western blot (Supp. Figure S1). Two out of three missense variants in the Estonian short extremes decreased NPR-B activity (c.2449G>A (p.E817K) and c.1982C>A (p.T661K)). Interestingly, one of two missense variants identified in FINRISK tall extremes showed increased NPR-B activity in both assays (c.1685G>A (p.R562Q)), strongly suggesting this is a gain of function variant contributing to the individuals’ tall stature. The second variant, which showed increased NPR-B activity only in the whole cell lysate assay, was also observed in Estonian short extreme samples, making it even less likely to be pathogenic.
Verifying the role of heterozygous functional NPR2 variants in ISS individuals
To test formally whether there are more loss of function NPR2 variants in short stature individuals than in controls without short stature, we combined observations from the three cohorts that have control samples (in other words, excluding the second clinical short stature cohort), considering validated variants seen either only in cases or only in controls. In total, there are six likely loss-of-function mutations (five missense and one splice site) based on segregation and functional evidence, all observed in short samples, and one gain-of-function mutation in tall samples. Assuming that all variants are equally likely to occur in short stature samples and tall extreme samples (or control samples of normal height) under the null hypothesis, this observation gives a p value of 0.008 (one-tailed test, p value is the probability of the observed outcome i.e. 6:0 plus 0:1 and all more extreme). The estimated prevalence of loss of function NPR2 variants in ISS individuals ranged from 1/26 to 0, and no variants were observed in the cohort of 136 short individuals from Finland (Table 6).
Table 6.
Fraction of ISS individuals with clearly functional NPR2 variants
| Cohort | # of functional NPR2 variants |
# of ISS individuals |
Fraction of ISS individuals with functional NPR2 variants |
|---|---|---|---|
| Short stature patients and FHS controls | 4 | 104 | 1/26 |
| Second short stature cohort | 2 | 216 | 1/108 |
| Extremes of FINRISK height distribution | 0 | 136 | 0 |
| Extremes of Estonian Biobank height distribution | 2 | 500 | 1/250 |
Discussion
To explore the role of NPR2 variation in short stature, we screened for nonsynonymous NPR2 variants in four different cohorts (Table 4). Familial segregation analyses in the short stature patient cohort supported the hypothesis that rare heterozygous NPR2 loss of function variants contribute strongly to short stature in individuals carrying these variants. We also identified rare nonsynonymous NPR2 variants in three additional cohorts, where family data are not available. Functional studies of the NPR2 missense variants identified eight loss-of-function mutations in short stature samples and one gain-of-function mutation in tall stature samples. The frequencies of heterozygous mutation carriers in ISS individuals varied across cohorts from 1/26 to 0, and no variants were observed in the short individuals from Finland. This variability may be due to different ancestries, variable sample selection criteria, and statistical fluctuation. Of note, for the second short stature cohort, coverage was not 100% for 5 out of 22 exons and exon 3 was not evaluated. This can reduce the observed frequency of allelic variants found in this group of patients.
In contrast with homozygous mutations in NPR2, which produces a severe short stature and body disproportion, heterozygous mutations in NPR2 seem to be associated with mild and variable growth impairment without a distinct skeletal phenotype. In our studies, the severity of short stature, body proportions, and presence of nonspecific skeletal abnormalities varied across individuals (Supp. Table S1), consistent with previous observations (Olney et al., 2006; Vasques et al., 2013). This variability is likely due to differences in the nature of NPR2 mutations carried by the individuals, as well as variable expressivity. Data was available on growth hormone response in three of our patients with pathogenic NPR2 variants and response was quite variable (Supp. Figures S2–S4).
Previously, a study of 47 Brazilian patients identified heterozygous NPR2 mutations in 6% of patients (Vasques et al., 2013), while another study on 101 short stature Japanese patients identified mutations in 2% of patients (Amano et al., 2014). Functional analyses were performed in both studies to evaluate the pathogenicity and elucidate the molecular mechanisms of the identified mutations. However, both studies included small numbers of patients and lacked the controls needed to formally show an enrichment of loss of function variants in ISS patients. Our study, including much larger cohorts, controls, family data, and functional analyses, is a more comprehensive assessment of heterozygous mutations in NPR2 as a potential cause of growth impairment in ISS patients.
Our study does show that different methods of analyzing the function of NPR2 variants can produce different results. The whole cell lysate method under the conditions specified is less sensitive at detecting partial loss of function mutations compared to a more rigorous enzyme based assay conducted on membrane fractions of transfected cells using known concentrations of substrate. However, the enzyme based assay is performed in crude membranes from broken cell preparations and cannot evaluate the ability of the mutations to affect the concentrations of the receptors that are properly targeted to the cell surface. Therefore, mutations that reduce activity in the enzyme assay are likely to be damaging but this assay may not detect mutations that reduce activity due to inappropriate plasma membrane targeting or mutations that affect receptor degradation.
The functional assays used do have limitations. Although the rat NPR2 construct is >98% identical to human NPR2 and identical at each of the amino acids mutated in this study, there remains the possibility that non-identical residues located near mutant residues could affect protein function. In addition, the assays do not provide mechanistic data for why these heterozygous mutations reduced NPR2 function. Neither assay is able to differentiate between loss-of-function due to haploinsuffiency and loss of function due to dominant negative effect of the mutant allele.
The frequency of heterozygous carriers of AMDM mutations was previously estimated to be about 0.14% (Olney et al., 2006). In the NHLBI Exome Sequencing Project, the cumulative frequency of all nonsynonymous NPR2 variants is approximately 0.4% in ~4000 European American samples. The discrepancy between these two allele frequencies highlights the importance of functional studies, which can help distinguish neutral background variants from true loss of function variants. In our study, out of 14 rare nonsynonymous NPR2 variants that were functionally tested, only 6 showed decreased functional activity. Although the variants with no in vitro functional consequences may still contribute to short stature, our in vitro findings are consistent with the 3-fold higher frequency of nonsynonymous variants in the population database compared with expectation based on estimated AMDM carrier rates.
Despite the rarity of AMDM mutations (and nonsynonymous NPR2 variants in general), these loss-of-function alleles likely have relatively large effect sizes (loss-of-function alleles were previously estimated to reduce height by ~1.8 SD (Olney et al., 2006)). Assuming an effect size of 1.8 SD and a cumulative population allele frequency of 0.14%, these maximum-likelihood estimates predict that ~2.6% of individuals with height below −2 SD would have a loss of function NPR2 variant and that ~4.2% of individuals with height below the 1st percentile would carry such a variant. These estimates are somewhat higher than our empirical findings in the two clinical short stature cohorts combined (1.9%), and also substantially higher than our estimates from the Finnish and Estonia cohorts (0.3% of individuals below the ~1st percentile), suggesting that the effect size or the population allele frequencies previously estimated for loss-of-function NPR2 variants are inflated. The higher reported rates of NPR2 mutations in previous studies (Vasques et al., 2013; Amano et al., 2014) could also be due to population differences, or more stringent selection criteria in earlier studies that made it more likely to ascertain individuals carrying NPR2 loss of function mutations (such as high rates of familial short stature). Interestingly, we also identified one gain-of-function NPR2 variant in tall extremes of FINRISK height distribution, suggesting NPR2 gain of function may be a more common cause of tall stature than previously recognized.
AMDM patients have been posited to have an abnormality in the GH/IGF-1 system, characterized by low insulin-like growth factor 1 (IGF-1; MIM# 147440) levels, high growth hormone (GH) levels, and lack of a response to GH treatment (Olney et al., 2006). However, IGF-1 levels were not low in either the carriers of AMDM mutations (Olney et al., 2006; Vasques et al., 2013; Amano et al., 2014) or in the subjects in our cohort with loss of function mutations for whom data were available. Larger prospective studies are needed to determine whether heterozygous carriers of clearly pathogenic NPR2 variants will respond to growth hormone. Regardless, NPR2 loss-of-function variants likely account for approximately 0.4–4% of all patients with short stature (height < −2 SD), and should be considered in the diagnostic evaluation of patients presenting with ISS. Interestingly, we identified NPR2 loss-of-function variants in 13.6% of familial cases, suggesting that such variants will provide a frequent explanation for patients with familial ISS.
Supplementary Material
Acknowledgments
Disclosure Statement: J.N.H. received a grant from Pfizer that did not fund any work described in this manuscript.
Grant Sponsor:
This work was conducted with support from Harvard Catalyst – The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Award 8UL1TR000170-05 and financial contributions from Harvard University and its affiliated academic health care centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, or the National Institutes of Health. Sequencing experiments were performed by the Sequencing Core Facility of the Molecular Genetics Core Facility at Boston Children’s Hospital supported by NIH P30-HD18655. This work was also supported by NIH Grants 1K23HD073351 (to A.D.), R01GM098309 (to L.R.P.), T32AR050938 (to J.W.R. and A.B.E.), March of Dimes grant 6-FY12-393 (to J.N.H.), funding from Genetech Center for Clinical Research in Endocrinology (to R.C.O.) and funding from the Howard Hughes Medical Institute (to MLW).
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano N, Mukai T, Ito Y, Narumi S, Tanaka T, Yokoya S, Ogata T, Hasegawa T. Identification and Functional Characterization of Two Novel NPR2 Mutations in Japanese Patients with Short Stature. J. Clin. Endocrinol. Metab. 2014:jc20133525. doi: 10.1210/jc.2013-3525. [DOI] [PubMed] [Google Scholar]
- Bartels CF, Bükülmez H, Padayatti P, Rhee DK, Ravenswaaij-Arts C, van Pauli RM, Mundlos S, Chitayat D, Shih L-Y, Al-Gazali LI, Kant S, Cole T, et al. Mutations in the transmembrane natriuretic peptide receptor NPR-B impair skeletal growth and cause acromesomelic dysplasia, type Maroteaux. Am. J. Hum. Genet. 2004;75:27–34. doi: 10.1086/422013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocciardi R, Giorda R, Buttgereit J, Gimelli S, Divizia MT, Beri S, Garofalo S, Tavella S, Lerone M, Zuffardi O, Bader M, Ravazzolo R, et al. Overexpression of the C-type natriuretic peptide (CNP) is associated with overgrowth and bone anomalies in an individual with balanced t(2;7) translocation. Hum. Mutat. 2007;28:724–731. doi: 10.1002/humu.20511. [DOI] [PubMed] [Google Scholar]
- Chusho H, Tamura N, Ogawa Y, Yasoda A, Suda M, Miyazawa T, Nakamura K, Nakao K, Kurihara T, Komatsu Y, Itoh H, Tanaka K, et al. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc. Natl. Acad. Sci. U. S. A. 2001;98:4016–4021. doi: 10.1073/pnas.071389098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannema SE, Duyvenvoorde HA, van Premsler T, Yang R-B, Mueller TD, Gassner B, Oberwinkler H, Roelfsema F, Santen GWE, Prickett T, Kant SG, Verkerk AJMH, et al. An activating mutation in the kinase homology domain of the natriuretic peptide receptor-2 causes extremely tall stature without skeletal deformities. J. Clin. Endocrinol. Metab. 2013;98:E1988–E1998. doi: 10.1210/jc.2013-2358. [DOI] [PubMed] [Google Scholar]
- Krejci P, Masri B, Fontaine V, Mekikian PB, Weis M, Prats H, Wilcox WR. Interaction of fibroblast growth factor and C-natriuretic peptide signaling in regulation of chondrocyte proliferation and extracellular matrix homeostasis. J. Cell Sci. 2005;118:5089–5100. doi: 10.1242/jcs.02618. [DOI] [PubMed] [Google Scholar]
- Kuczmarski RJ, Ogden CL, Guo SS, Grummer-Strawn LM, Flegal KM, Mei Z, Wei R, Curtin LR, Roche AF, Johnson CL. 2000 CDC Growth Charts for the United States: methods and development. Vital Health Stat. 2002;11:1–190. [PubMed] [Google Scholar]
- Leitsalu L, Haller T, Esko T, Tammesoo M-L, Alavere H, Snieder H, Perola M, Ng PC, Mägi R, Milani L, Fischer K, Metspalu A. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol. 2014 doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- Mericq V, Uyeda JA, Barnes KM, Luca F, De, Baron J. Regulation of fetal rat bone growth by C-type natriuretic peptide and cGMP. Pediatr. Res. 2000;47:189–193. doi: 10.1203/00006450-200002000-00007. [DOI] [PubMed] [Google Scholar]
- Miura K, Kim O-H, Lee HR, Namba N, Michigami T, Yoo WJ, Choi IH, Ozono K, Cho T-J. Overgrowth syndrome associated with a gain-of-function mutation of the natriuretic peptide receptor 2 (NPR2) gene. Am. J. Med. Genet. A. 2014;164A:156–163. doi: 10.1002/ajmg.a.36218. [DOI] [PubMed] [Google Scholar]
- Miura K, Namba N, Fujiwara M, Ohata Y, Ishida H, Kitaoka T, Kubota T, Hirai H, Higuchi C, Tsumaki N, Yoshikawa H, Sakai N, et al. An overgrowth disorder associated with excessive production of cGMP due to a gain-of-function mutation of the natriuretic peptide receptor 2 gene. PLoS One. 2012;7:e42180. doi: 10.1371/journal.pone.0042180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncla A, Missirian C, Cacciagli P, Balzamo E, Legeai-Mallet L, Jouve J-L, Chabrol B, Merrer M, Le, Plessis G, Villard L, Philip N. A cluster of translocation breakpoints in 2q37 is associated with overexpression of NPPC in patients with a similar overgrowth phenotype. Hum. Mutat. 2007;28:1183–1188. doi: 10.1002/humu.20611. [DOI] [PubMed] [Google Scholar]
- National Heart, Lung and BI [Google Scholar]
- Olney RC, Bükülmez H, Bartels CF, Prickett TCR, Espiner EA, Potter LR, Warman ML. Heterozygous mutations in natriuretic peptide receptor-B (NPR2) are associated with short stature. J. Clin. Endocrinol. Metab. 2006;91:1229–1232. doi: 10.1210/jc.2005-1949. [DOI] [PubMed] [Google Scholar]
- Potter LR, Abbey-Hosch S, Dickey DM. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr. Rev. 2006;27:47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, Boucher G, Ripke S, Ellinghaus D, Burtt N, Fennell T, Kirby A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JW, Dickey DM, Miura K, Michigami T, Ozono K, Potter LR. A human skeletal overgrowth mutation increases maximal velocity and blocks desensitization of guanylyl cyclase-B. Bone. 2013;56:375–382. doi: 10.1016/j.bone.2013.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JW, Potter LR. ATP potentiates competitive inhibition of guanylyl cyclase A and B by the staurosporine analog, Gö6976: reciprocal regulation of ATP and GTP binding. J. Biol. Chem. 2011;286:33841–33844. doi: 10.1074/jbc.M111.273565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock MJ, Prenen J, Funari VA, Funari TL, Merriman B, Nelson SF, Lachman RS, Wilcox WR, Reyno S, Quadrelli R, Vaglio A, Owsianik G, et al. Gain-of-function mutations in TRPV4 cause autosomal dominant brachyolmia. Nat. Genet. 2008;40:999–1003. doi: 10.1038/ng.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S. C-type natriuretic peptide and guanylyl cyclase B receptor. Peptides. 2005;26:1024–1034. doi: 10.1016/j.peptides.2004.08.027. [DOI] [PubMed] [Google Scholar]
- Tamura N, Doolittle LK, Hammer RE, Shelton JM, Richardson JA, Garbers DL. Critical roles of the guanylyl cyclase B receptor in endochondral ossification and development of female reproductive organs. Proc. Natl. Acad. Sci. U. S. A. 2004;101:17300–17305. doi: 10.1073/pnas.0407894101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen E, Laatikainen T, Peltonen M, Juolevi A, Männistö S, Sundvall J, Jousilahti P, Salomaa V, Valsta L, Puska P. Thirty-five-year trends in cardiovascular risk factors in Finland. Int. J. Epidemiol. 2010;39:504–518. doi: 10.1093/ije/dyp330. [DOI] [PubMed] [Google Scholar]
- Vasques GA, Amano N, Docko AJ, Funari MFA, Quedas EPS, Nishi MY, Arnhold IJP, Hasegawa T, Jorge AAL. Heterozygous Mutations in Natriuretic Peptide Receptor-B (NPR2) Gene as a Cause of Short Stature in Patients Initially Classified as Idiopathic Short Stature. J. Clin. Endocrinol. Metab. 2013;98:E1636–E1644. doi: 10.1210/jc.2013-2142. [DOI] [PubMed] [Google Scholar]
- Wang SR, Carmichael H, Andrew SF, Miller TC, Moon JE, Derr MA, Hwa V, Hirschhorn JN, Dauber A. Large Scale Pooled Next-Generation Sequencing of 1077 Genes To Identify Genetic Causes of Short Stature. J. Clin. Endocrinol. Metab. 2013 doi: 10.1210/jc.2013-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasoda A, Komatsu Y, Chusho H, Miyazawa T, Ozasa A, Miura M, Kurihara T, Rogi T, Tanaka S, Suda M, Tamura N, Ogawa Y, et al. Overexpression of CNP in chondrocytes rescues achondroplasia through a MAPK-dependent pathway. Nat. Med. 2004;10:80–86. doi: 10.1038/nm971. [DOI] [PubMed] [Google Scholar]
- Yoder AR, Kruse AC, Earhart CA, Ohlendorf DH, Potter LR. Reduced ability of C-type natriuretic peptide (CNP) to activate natriuretic peptide receptor B (NPR-B) causes dwarfism in lbab −/− mice. Peptides. 2008;29:1575–1581. doi: 10.1016/j.peptides.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.