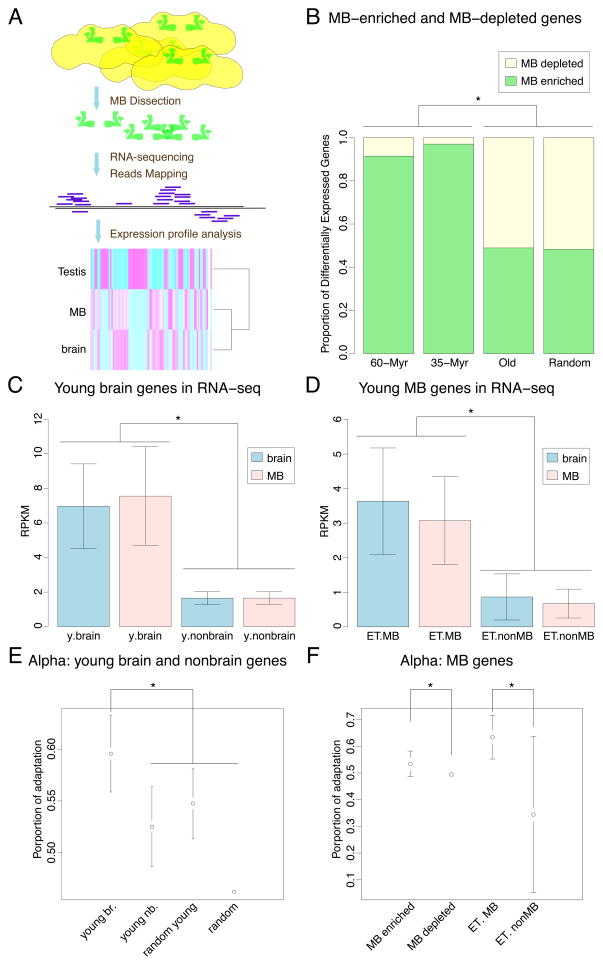
Figure 3. Adaptive evolution of young brain and young MB genes.
(A) Schematic representation of RNA-seq showing the workflow of experimental (dissection, RNA-sequencing) and analytic procedures, with a heatmap of young gene expression in MB, brain and testis at the bottom.
(B) Statistical summary of MB-enriched and MB-depleted genes in differentially expressed gene sets; 60-Myr, genes younger than 60 million years (n=447, DE=58); 35-Myr, genes younger than 35 million years (n=279, DE=33); Old, genes older than 60 Myr (n=9275, DE=2272); Random, randomly sampled genes from the genome (n=11820, DE=2940); DE, number of differentially expressed genes.
(See also Figure S3B)
(C) Relative gene expression abundance (mean RPKM values from biological replicates) of young brain genes (y.brain) and young nonbrain genes (y.nonbrain) detected by RT-PCR, in brain and MB RNA-seq datasets.
(D) Relative gene expression abundance of young MB genes (ET.MB) and young genes not expressed in MB (ET.nonMB) detected by enhancer trap, in brain and MB RNA-seq datasets.
(E) Estimation of Alpha, the proportion of nonsynonymous substitutions subjected to positive selection, revealed positive selection on young brain genes. Young brain genes (young br.) have significantly higher alpha compared to young nonbrain genes (young nb.), random young genes (random young) or random genes (random) representing the genomic background from random sampling. The “random” error bar is too small to display.
(F) Estimation of Alpha revealed positive selection on MB genes. MB-enriched genes have significantly higher alpha compared to MB-depleted genes; young MB genes (ET. MB) have significantly higher alpha compared to young nonMB genes (ET. nonMB). The “MB-depleted” error bar is too small to display. Stars denote significance in statistical comparison, if applicable.
(See also: Figure S3)