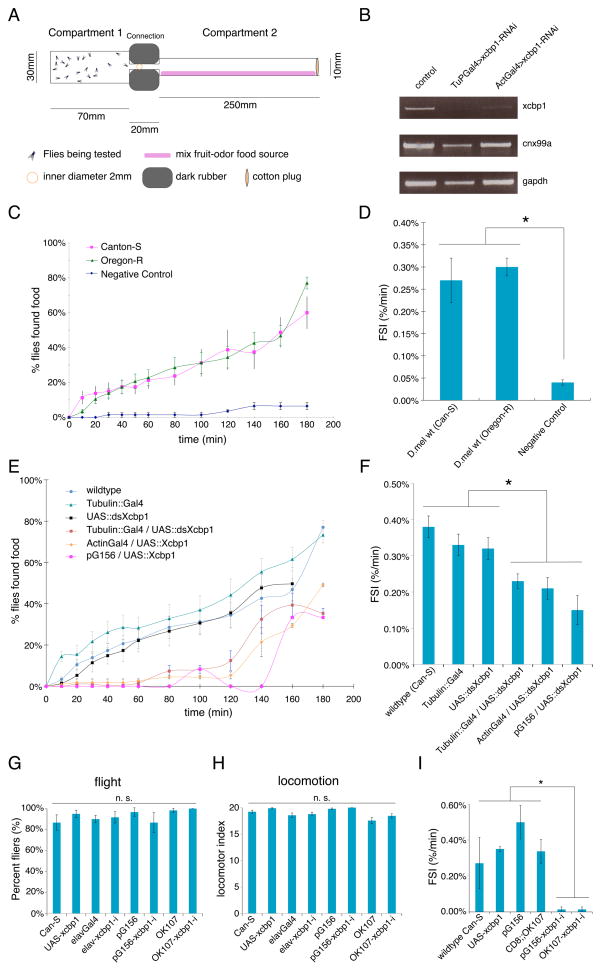
Figure 5. Xcbp1 influences foraging behavior.
(A) Scheme of the foraging assay system design.
(B) Semi-quantitative RT-PCR showing the specific and efficient RNAi knockdown of Xcbp1. Sample genotypes are shown above and assayed genes on the right.
(C) Foraging curve of wt D. melanogaster strains showing the time-dependent increase in the percentage of flies that found food. Flies do not crawl to Compartment 2 if the food source is not present (negative control).
(D) Foraging speed index (FSI) measurements of wt D. melanogaster strains.
(E) Foraging curve for Xcbp1 RNAi in D. melanogaster with constitutive Gal4s and Xcbp1-Gal4 (pG156) or in control (wt, Gal4 only, UAS only) animals. Genotypes are shown in the inset legend.
(F) FSI measurements of control or Xcbp1 RNAi knockdown animals. Loss of Xcbp1 results in a significant decrease in foraging ability. Genotypes are shown at the bottom of each column.
(G–H) Xcbp1 RNAi knockdown animals show no significant impairment of either flight (G) or locomotion (H) when compared to controls. Genotypes are shown at the bottom of each column.
(I) FSI measurements of control or Xcbp1 MB RNAi knockdown animals. Loss of Xcbp1 in MB results in a significant decrease in foraging ability. Genotypes are shown at the bottom of each column.
In barplots, data are represented as mean +/− SEM (standard error of the mean), where error bars denote SEM values; asterisks denote statistical significance (p < 0.01 unless otherwise noted) in all pairwise comparisons; n.s., not significant (p > 0.01 unless otherwise noted) in all pairwise comparisons.
(See also: Figure S5)