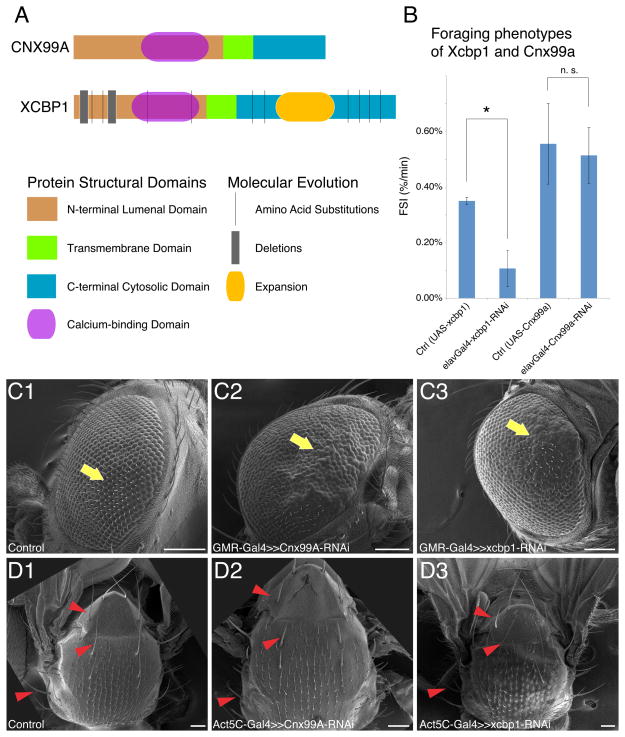
Figure 6. Conservation and divergence of the Xcbp1-Cnx99a gene pair.
(A) Scheme of the XCBP1 and CNX99A proteins. N-terminal domains (NTDs), brown; transmembrane domain (TM), green; C-terminal domains (CTDs), cyan. Amino acid substitutions, deletions and expansions are noted on the Xcbp1 diagram.
(B) FSI measurements of control, Xcbp1 RNAi knockdown and Cnx99a RNAi knockdown animals. Genotypes are shown at the bottom of each column. Data are represented as mean +/− SEM. The asterisk denotes statistical significance (p < 0.01); n.s., not significant (p > 0.01).
(C) SEM images of eye morphology of control animals (C1) or animals with Cnx99a (C2) or Xcbp1 (C3) RNAi knockdown. Yellow arrows point to normal ommatidial structures in control, and defective or overproliferated ommatidial structures in both Cnx99a and Xcbp1 RNAi animals. Scale bars = 100 μm
(D) SEM images of notum morphology of control animals (D1) or animals with Cnx99a (D2) or Xcbp1 (D3) RNAi knockdown. Red arrowheads denote shorter and blunter bristles in Cnx99a RNAi that are unaffected in control or Xcbp1 RNAi flies. Scale bars = 100 μm.
(See also: Figure S6)