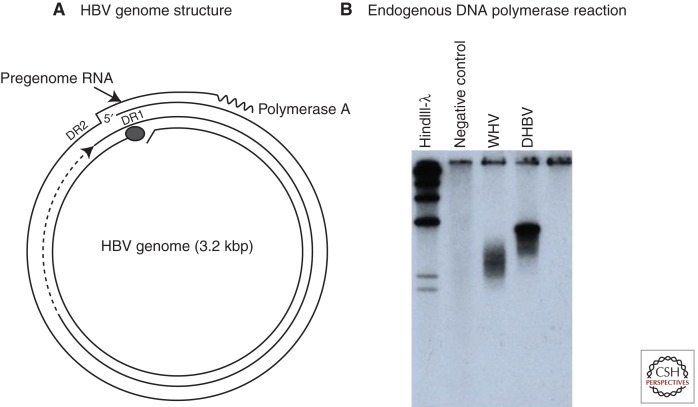
Figure 1.
The genome and endogenous polymerase reaction of hepatitis B virus (HBV). (A) The HBV genome is a partially double-stranded DNA, held in a relaxed circular conformation by a short cohesive overlap between the 5′ ends of the two DNA strands (Summers et al. 1975). One strand, later found to be the plus strand, is always incomplete in virus particles, with a gap that may encompass up to 50% of the genome length. The minus strand is always complete. The large circle at the 5′ end of the minus strand represents a covalently attached protein (Gerlich and Robinson 1980) that was later shown to be the viral DNA polymerase/reverse transcriptase (Bartenschlager and Schaller 1988). Pregenomic RNA, the template for viral DNA synthesis, is shown for comparison. DR1 and DR2 are 11-nucleotide direct repeats (12-nucleotide for duck hepatitis B virus [DHBV]) on the pregenome that play essential roles in priming of viral DNA synthesis (see text). (B) HBV and other hepadnaviruses contain the viral DNA polymerase, which can fill in the single-stranded gap in vitro (Summers et al. 1975). The fill-in reaction can be performed by pelleting virus from serum, adding nonionic detergent and radiolabeled deoxynucleotides (dNTPs) to the pellet, and incubating at 37°C. The radiolabeled DNA can be detected by agarose gel electrophoresis and autoradiography, as shown here for DHBV and woodchuck hepatitis virus (WHV). This assay was instrumental in the discovery of WHV, DHBV, and ground squirrel hepatitis virus (GSHV) (Summers et al. 1978; Marion et al. 1980; Mason et al. 1980).