Figure 3.
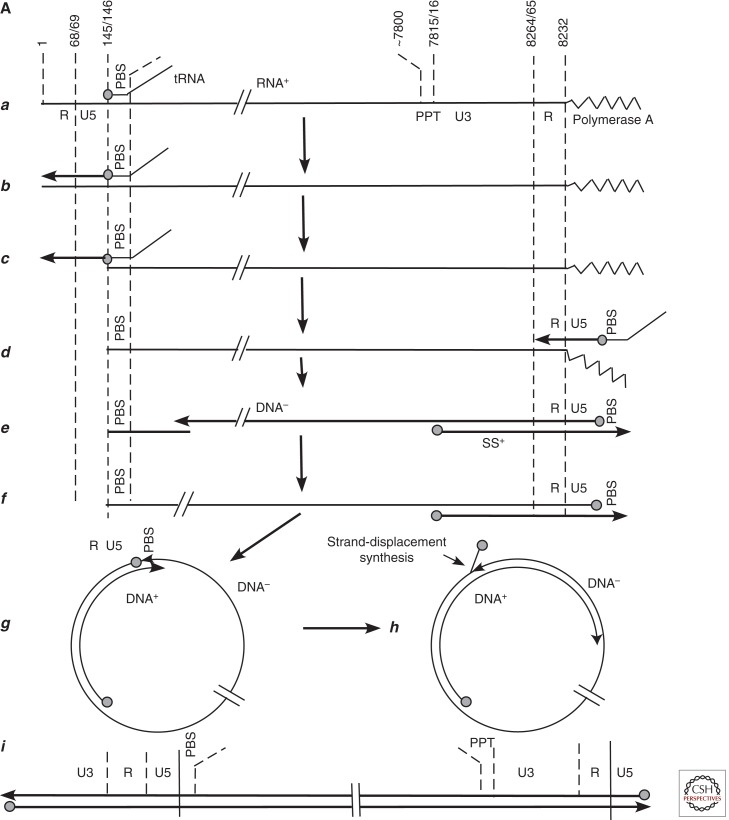
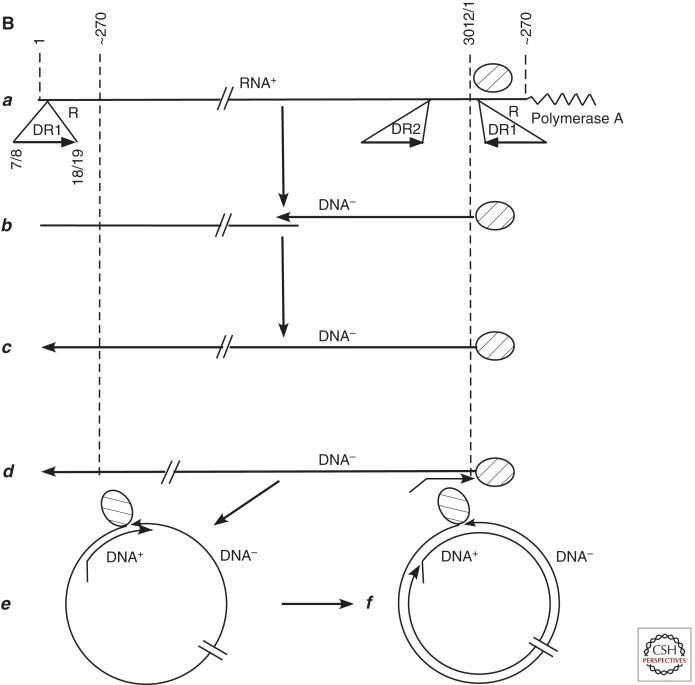
Comparison of retrovirus and hepadnavirus DNA synthesis pathways. (A) Moloney murine leukemia virus (MoMLV) DNA synthesis. Reverse transcription of MoMLV RNA begins when a cell is infected. The primer for reverse transcription is the 3′ hydroxyl (OH) of tRNAPro, which is annealed near to the 5′ end of the viral RNA genome via hybridization to an 18-nucleotide primer binding site (PBS) (a). Synthesis extends to the 5′ end of the viral RNA. The reverse-transcribed RNA sequences are degraded by the viral RNase H (b,c). At this point, the DNA product can hybridize to the 3′ end of viral RNA through the 68-nucleotide R domain, found at both ends of viral RNA (d). This facilitates reverse transcription of the remainder of the RNA template. Plus-strand synthesis initiates near the 5′ end of the minus strand from an RNA oligonucleotide that is left behind during RNase H degradation of the reverse-transcribed viral RNA (e). (It should be noted that some retroviruses prime plus-strand synthesis from multiple sites, including the polypurine tract [PPT] region as well as upstream sites.) Plus-strand synthesis extends rightward to copy the 3′ 18 nucleotides of the tRNA, recreating the PBS as DNA. Once minus-strand synthesis has made a complementary copy of the PBS (f), the 3′ ends of the nascent plus and minus strands can hybridize to form a circle (g). Minus-strand synthesis extends to the 5′ end of the plus strand, presumably via strand displacement synthesis (h), and plus-strand synthesis extends to the 5′ end of the minus strand to create a DNA with a large terminal redundancy (LTR; U3-R-U5). This terminally redundant linear DNA is the substrate for integration into host DNA. Integrated DNA serves as the provirus template for new viral RNA synthesis. (B) duck hepatitis B virus (DHBV) DNA synthesis. In this early model (Lien et al. 1986), reverse transcription is primed from a tyrosine residue of a protein primer, later shown to map to the terminal protein domain of the viral DNA polymerase (a), and begins within the 3′ copy of DR1. Minus-strand synthesis extends to the 5′ end of the pregenome (b,c). Because it began in R and extended almost to the 5′ end of the RNA template, the nascent minus strand is terminally redundant by 7–8 nucleotides. The pregenome is degraded by RNase H, leaving an 18-nucleotide sequence including the 5′ CAP and first 18 nucleotides of the pregenome (c), including the 12-base repeat, DR1, which is present in the terminal redundancy of the pregenome. This oligoribonucleotide is then translocated from DR1 to DR2, which maps upstream of the 3′ terminal redundancy (d). The oligoribonucleotide hybridizes to DR2 on the minus-strand DNA because DR1 and DR2 are identical in sequence. Plus-strand synthesis initiates at the 3′ end of DR2 and extends to the end of the minus strand. Circularization to continue plus-strand synthesis is apparently facilitated by the 7- to 8-nucleotide terminal redundancy on the minus strand (e,f). Synthesis continues, to produce a partially double-stranded virus genome (with DHBV, most plus strands are nearly full-length, excluding DR2, to which the plus-strand primer remains bound). Approximately 10% of the time, the plus-strand primer is not translocated to DR2, and plus-strand synthesis begins at the upstream copy of DR1, resulting in a linear virus genome (see Fig. 4).