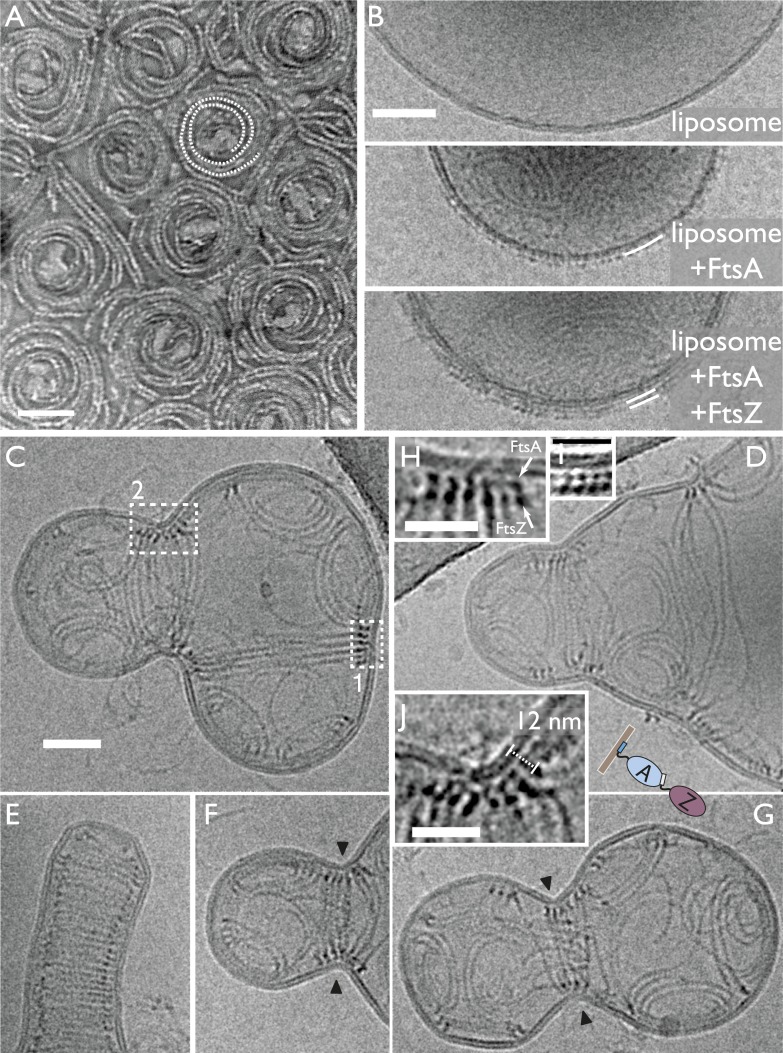
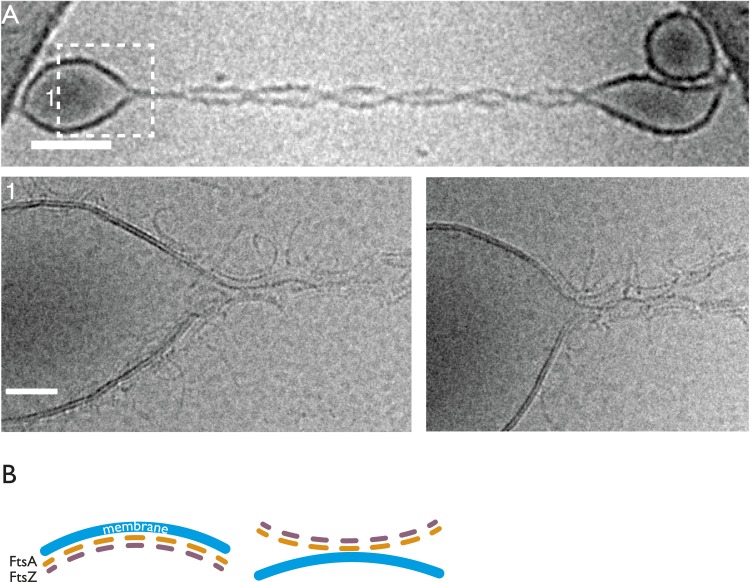
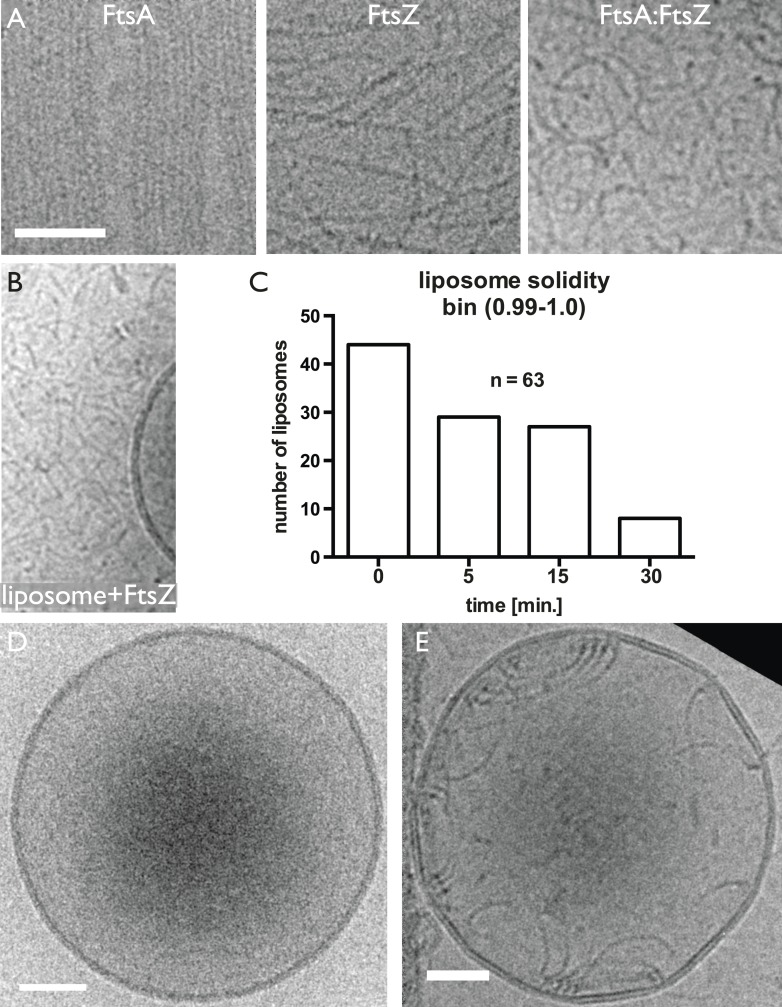
Figure 3. In vitro reconstitution of bacterial cell membrane constriction by the FtsZ ring from purified components.
(A) Thermotoga maritima FtsA (TmFtsA) and Thermotoga maritima FtsZ (TmFtsZ) form spirals on a flat lipid monolayer, as indicated by a white dotted line. The filaments tend to appear as double strands (doublets). Negative-stain electron microscopy. (B) Transmission electron cryomicroscopy allows resolution of the inner and outer leaflet of undisturbed liposomes (top panel). When TmFtsA is added to the outside, an additional layer of density corresponding to FtsA becomes apparent (middle panel). Recruitment of TmFtsZ by TmFtsA leads to the formation of two layers (bottom panel). Taken together, we conclude that FtsA is sandwiched between the membrane and FtsZ filaments (bottom panel). See also Figure 3—figure supplement 1 and Figure 3—figure supplement 2. (C–G) Constriction sites are efficiently formed when TmFtsA and TmFtsZ are encapsulated in liposomes that have sizes comparable to bacterial cells. Five representative liposomes are shown using transmission electron cryomicroscopy (hence are 2D projections of 3D objects). Importantly, constriction sites are only formed where a ring made of the two proteins is present (black arrowheads) and not at other sites where filaments are located. The TmFtsA and TmFtsZ layers are clearly visible (inset H, same as boxed area ‘1’ in C; inset J, same as boxed area ‘2’ in C and inset I, which is from Figure 4 electron cryotomography data) and the protein's organisation mirrors that present in E. coli cells (compare with Figure 2C). The distance of 12 nm between TmFtsZ and the membrane (inset J) resembles that found in over-expressing cells (see Figure 2G and also Figure 5C). (E) Intriguingly, liposomes are being constricted (partially) in the absence of added nucleotide. Scale bars: 50 nm in (A–C), 25 nm for insets.