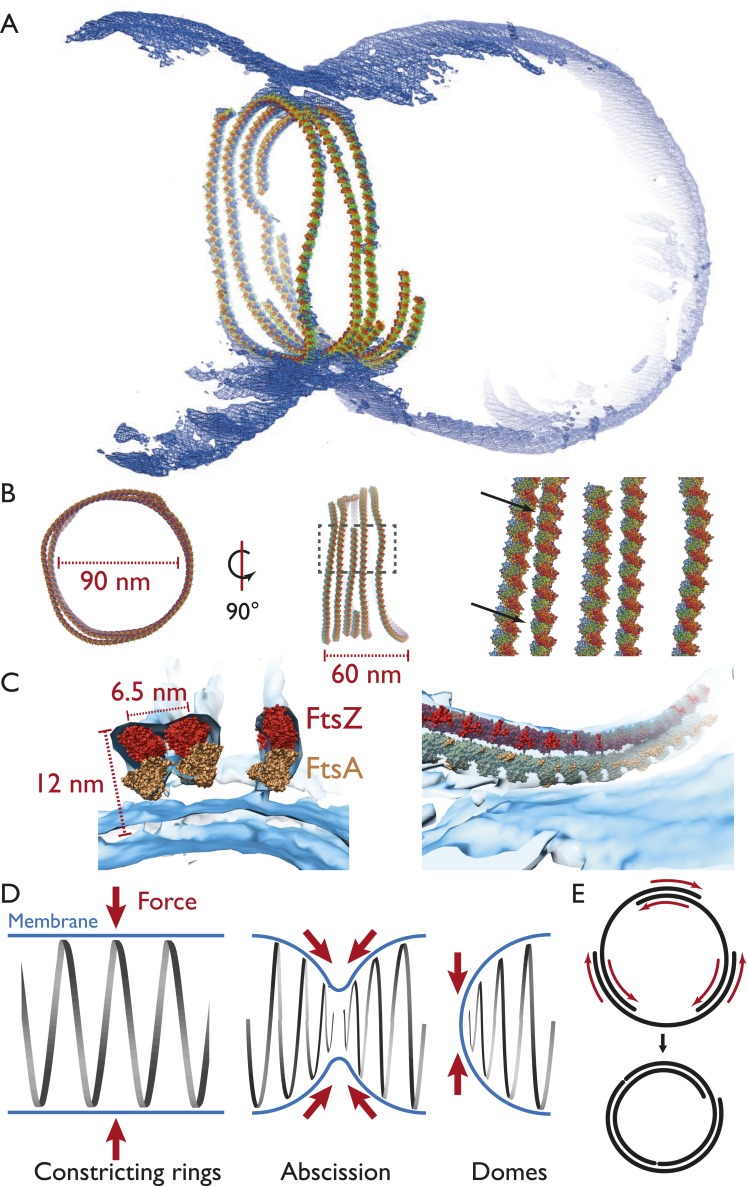
Figure 5. Visualising the FtsZ ring at the molecular level.
(A) A semi-atomic model of the FtsZ ring constricting a liposome. 294 monomers of S. aureus FtsZ have been roughly positioned using a spline-fitting approach (PDB 3VO8 (Matsui et al., 2012)). This uses the same tomography data as Figure 4A. (B) The ring is 90 nm in diameter (left) and 60-nm thick (middle). It consists of at least four individual filaments (right, atoms shown as spheres) with varying lateral interfilament distances (right, atoms shown as spheres, black arrows). (C) FtsZ filaments are single protofilaments, but they tend to pair in doublets. A precision manual fit of the TmFtsA polymer crystal structure (PDB 4A2B) (Szwedziak et al., 2012) in addition to 3VO8 FtsZ polymer crystal structure was performed in a region of very good density. The fit is excellent and dimensions and distances match well with CcFtsZ, EcFtsZ, and TmFtsAZ in vivo situations (Figure 1A,E, 2E,G). (D) Left: in the ring-like structures (black), force (red arrows) is perpendicular to the membrane (blue), leading to constriction. Middle: during constriction, the ring develops into two helical spirals, leading to forces pushing membrane inwards, and this might explain how abscission is accomplished since membranes will presumably not fuse while the protein filaments are in between (see Figure 4A bottom right and Video 9 for an example of this in liposomes). Right: the domes we observed do not deform liposomes because the force generated is almost perfectly tangential to the membrane. (E) Constriction force generation and filament sliding. In the discussion, three different energy sources for constriction are listed: maximising filament overlap, repeat mismatch within FtsA–FtsZ copolymers (Figure 4—figure supplement 2) and filament shortening and turnover due to nucleotide hydrolysis by FtsAZ. While it is currently not obvious which of these or if a combination of the three mechanisms drives constriction, it seems clear to us that constriction, at least in the liposome reconstitution experiments, requires filaments to slide past each other as is depicted in two dimensions. Since also unmodified wild-type cells (Figure 1) show closed continuous rings at division sites, we would assume the same holds true in vivo. Filament sliding can also explain the spirals on lipid monolayers (Figure 3A) and spirals in the dome-like structures with liposomes (Figure 4A). The schematic drawn is a simplification into two dimensions, of course, in vivo and in vitro FtsZ filaments overlap in the third dimension, forming single-layered bands since each filament is anchored to the membrane.