Fig. 3. Structural analysis of the 2-pyridone binding site in the InhA-NADH complex.
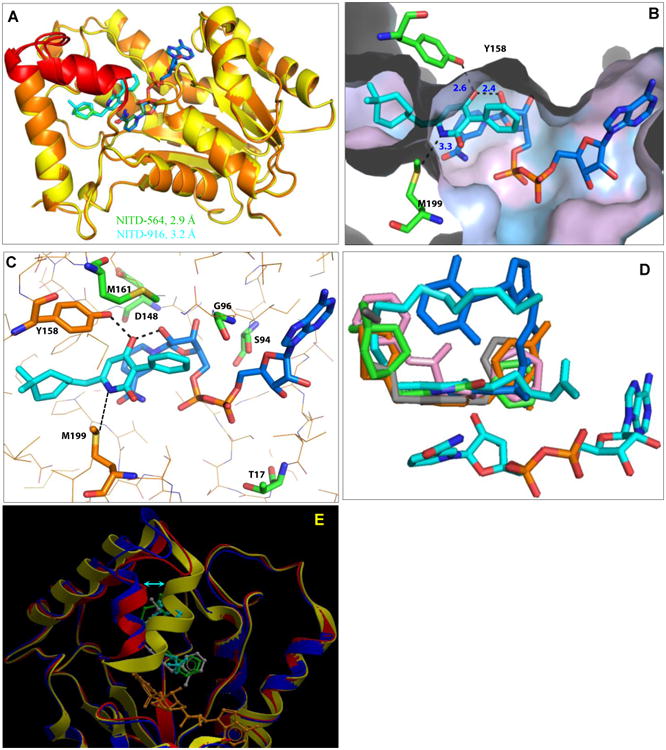
(A) Superimposed crystal structures of InhA-NADH-NITD564 (yellow) and InhA-NADH-NITD916 (orange); respective 2-pyridone ligands are shown in green and cyan. Substrate binding loop encompassing residues 196 to 211 is shown in red. (B) Close-up of NITD-916 (cyan) binding pocket in InhA-NADH complex, with protein polar (cyan) and hydrophobic (grey) surfaces shown. The side chains of Y158 and M199 residues are shown. The distance (in Å) between the ligand and side chains of with Y158, M199 and 2′-OH on the ribose sugar of NADH is highlighted by dotted lines. (C) Hydrogen bonding interactions of NITD-916 with critical residues in the active site of InhA. Side chains of amino acid residues responsible for NITD-916 resistance (T17, S94, G96, D148 and M161) are shown. (D) The InhA-NITD916 (green) structure overlaid with the fatty acyl substrate (cyan, 1BVR), along with other direct InhA inhibitors namely, triclosan derivative (orange, 3FNG), alkyl diphenyl ether (grey, 2×23), pyrrolidine carboxamides (pink, 2H7I) and methyl-thiazoles (blue, 4BQP). (E) The InhA-NITD916 structure (red) overlaid with co-crystal structures of fatty acyl substrate (blue, 1BVR) and alkyl diphenyl ether (yellow, 2×23). NITD-916, fatty acyl substrate and alkyl diphenyl ether ligands are colored in green, cyan and grey respectively. The shift in the conformation of the substrate binding loop is shown by an arrow.
