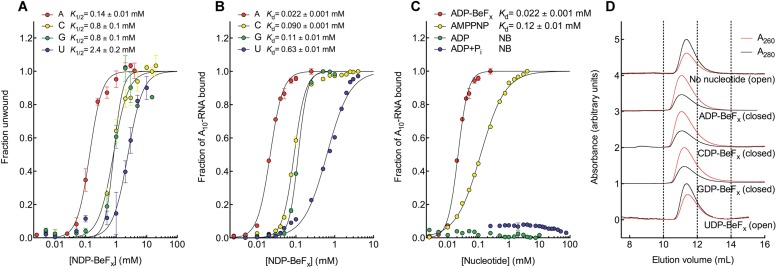
Figure 2. The biochemical basis for the ATP specificity of the helicase core of Mss116.
(A) dsRNA unwinding by the MBP-tagged helicase core measured under equilibrium conditions using a gel-based fluorescence assay to monitor the formation of a closed-state complex containing bound ssRNA at increasing concentrations of NDP-BeFx, N = A, C, G, or U (Figure 2—figure supplement 1). The fraction of unwound duplex was obtained by normalizing the band intensities separately for each gel using the parameters from the fit to a one-site binding model, as the change in fluorescence upon unwinding is different under each condition. The extent of unwinding with UDP-BeFx was less than that for the other nucleotide analogs, and the maximum concentration of UDP-BeFx used in this assay was insufficient to drive unwinding to completion (Figure 2—figure supplement 1). This could be because UDP-BeFx bound at saturating concentrations to D1 cannot efficiently induce a closed state. (B) Equilibrium binding of A10-RNA to the MBP-tagged helicase core determined by fluorescence anisotropy measurements at increasing concentrations of NDP-BeFx, N = A, C, G, or U. (C) Equilibrium binding of A10-RNA to the MBP-tagged helicase core determined as in (B) at increasing concentrations of ADP-BeFx, AMP-PNP, ADP, and ADP + Pi. Error bars in (A–C) represent the standard error for at least three independent measurements, and the error in the K1/2 or Kd represents the standard error of the non-linear regression. NB, no appreciable binding. In (B and C), the fraction of A10-RNA bound was calculated by normalizing against the anisotropy signal for unbound and fully bound substrate obtained from the fit to a one-site binding model. (D) Normalized SEC profiles monitored by absorbance at 260 nm (red) and 280 nm (black) for the helicase core in the absence of all substrates and in the presence of A10-RNA + NDP-BeFx, N = A, C, G, or U. An A260/A280 >1 at the maximum absorbance indicates the formation of a closed-state complex.