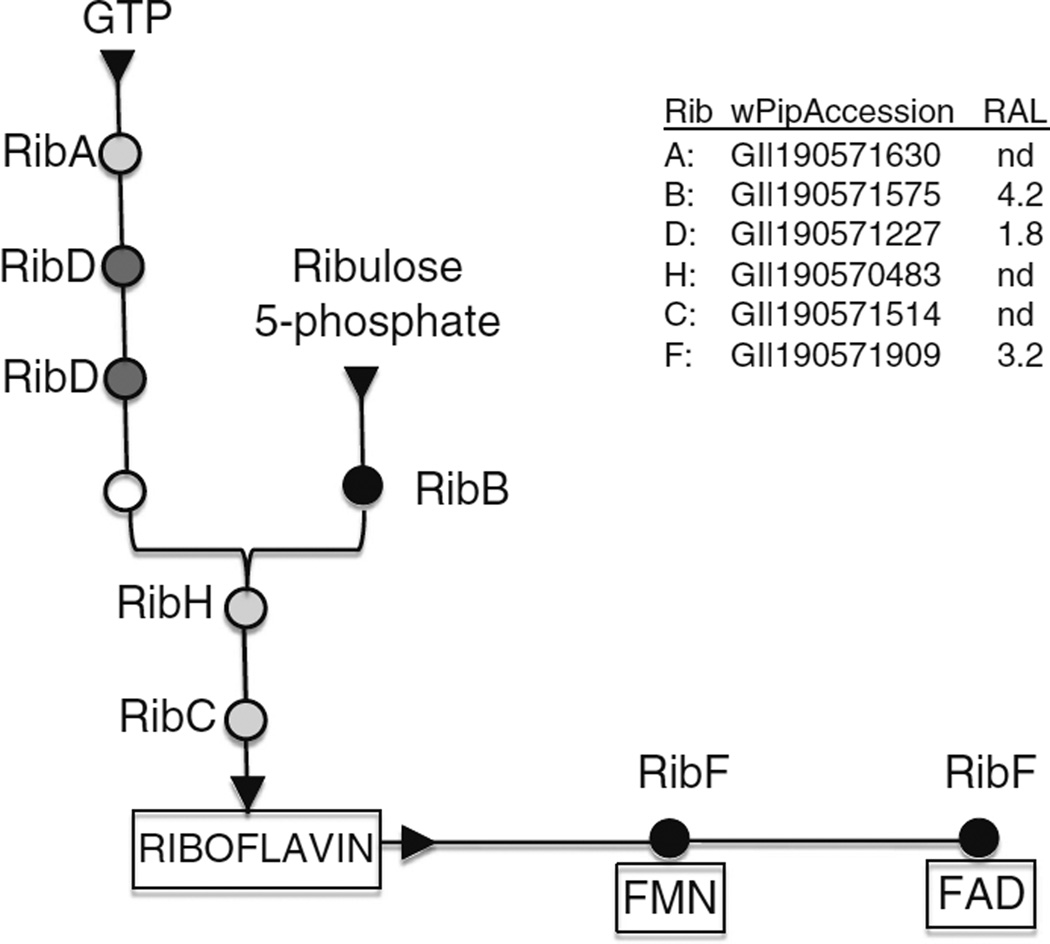
Figure 5.
Pathways for riboflavin biosynthesis in Wolbachia. Circles indicate enzymes that participate in riboflavin biosynthesis from two substrates, GTP derived from purine biosynthesis, and ribulose 5-phosphate derived from the pentose phosphate pathway. RibA GTP cyclohydrolase II; RibD bifunctional deaminase and reductase; open circle unidentified enzyme (see Bacher et al. 2000; Abbas and Sibirny 2011); RibB 3,4-dihydroxy-2-butanone 4-phosphate synthase; RibH lumazine synthase; RibC riboflavin synthase; RibF riboflavin kinase and FAD synthase. At the left, shading represents relative expression level for these pathways, based on data in a related proteomics study (Baldridge GD, Witthuhn BA, Higgins LA, Markowski TW, Fallon AM. Proteomic analysis of an insect Wolbachia in mosquito cells reveals robust expression of a constitutive stress response, a complete type IV secretion system and potential supplementation of bacterial amino acid pools by proline, glutamate, glycine and cysteine degradation, unpublished; see the legend at the right). Black represents highly expressed enzyme; dark gray represents moderately expressed; light gray represents not detected (nd) in the proteomics study. Accession numbers are from the annotated wPip (Pel) genome.