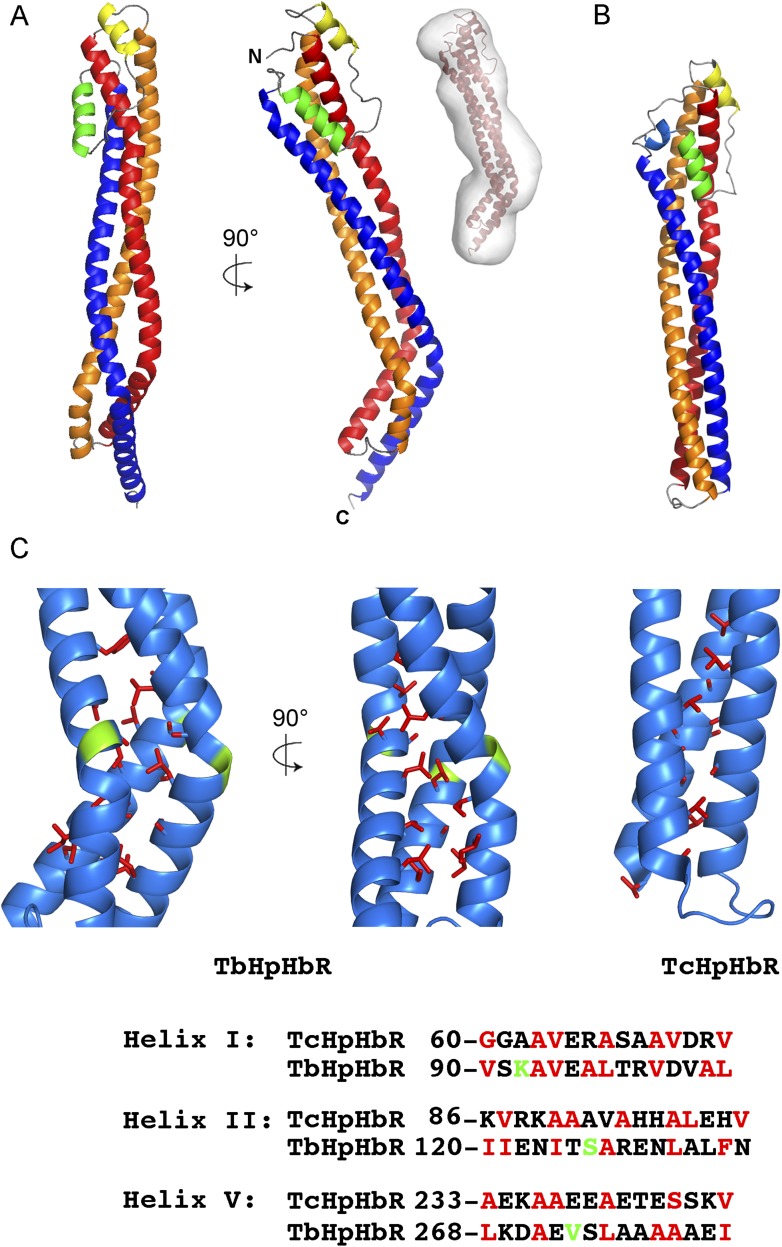
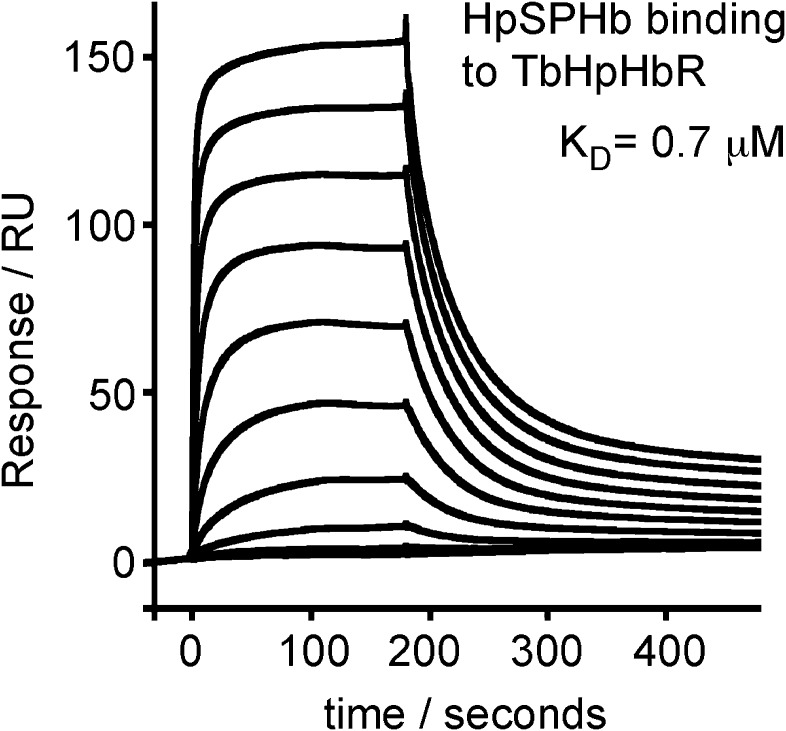
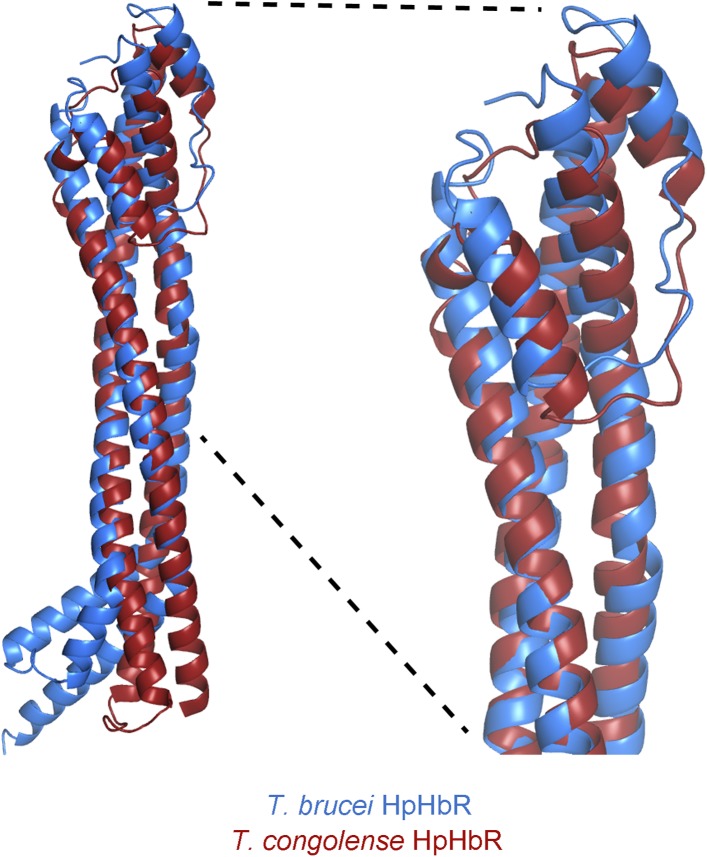
Figure 1. The structure of the T. brucei haptoglobin-haemoglobin receptor.
(A) The structure of the T. brucei haptoglobin-haemoglobin receptor, with helix I (red), helix II (orange) and helix V (blue). These three helices form an elongated bundle with a ∼50° kink towards the membrane proximal C-terminal end. The inset shows a molecular envelope derived from small angle x-ray scattering. (B) The structure of the T. congolense haptoglobin-haemoglobin receptor (Higgins et al., 2013) for comparison. (C) A change in the pattern of hydrophobic residues results in a rigid kink in the three helical bundle of the TbHpHbR. Corresponding regions of the structures of TbHpHbR and TcHpHbR are shown with side chains of the hydrophobic residues that pack in the core of the bundle coloured red and residues at the kink sites in TbHpHbR coloured green. Also shown are sequence alignments of TbHpHbR and TcHpHbR for these regions of each helix, coloured in the same way.