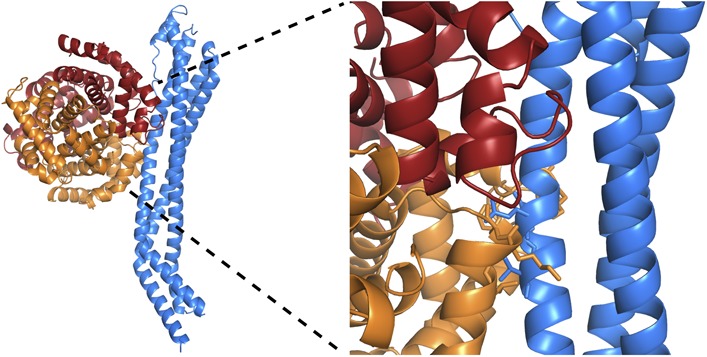
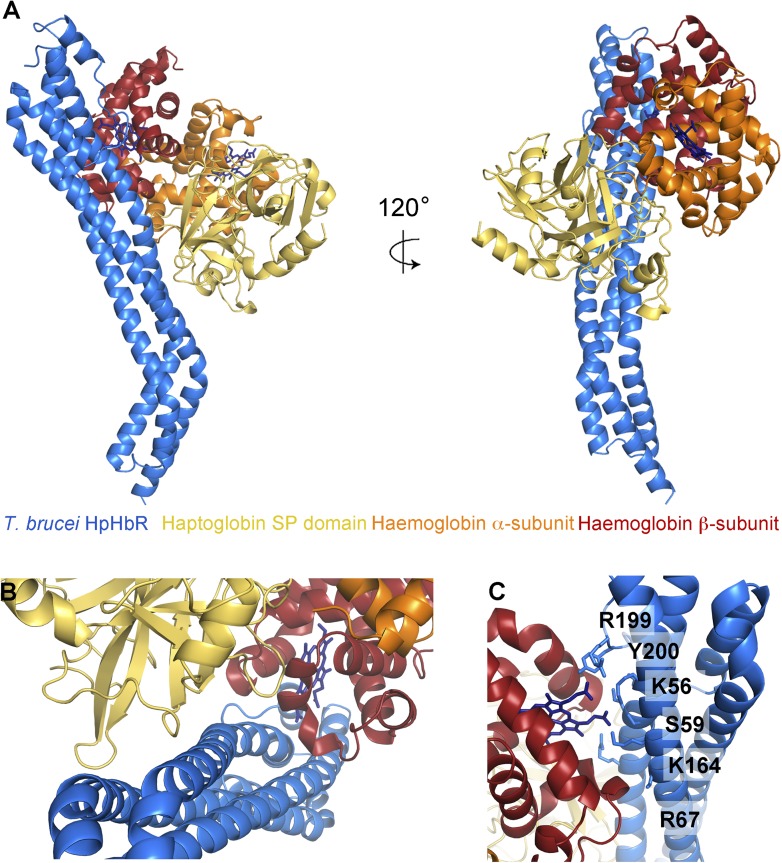
Figure 2. The structural basis for haptoglobin-haemoglobin binding by TbHpHbR.
(A) The structure of the complex between T. brucei HpHbR (blue) bound to its ligand, HpSPHb (haptoglobin is yellow, the β-subunit of haemoglobin is red and the α-subunit of haemoglobin is orange). (B) The complex viewed from the membrane proximal end, showing the contacts made by haptoglobin and the β-subunit of haemoglobin. (C) A view of the haemoglobin-binding site showing direct contacts between the haem and the receptor. Residues from the receptor that directly contact the haemoglobin subunit are shown as sticks and are numbered.
Figure 2—figure supplement 1. Stereoview of the TbHpHbR in complex with HpHb.
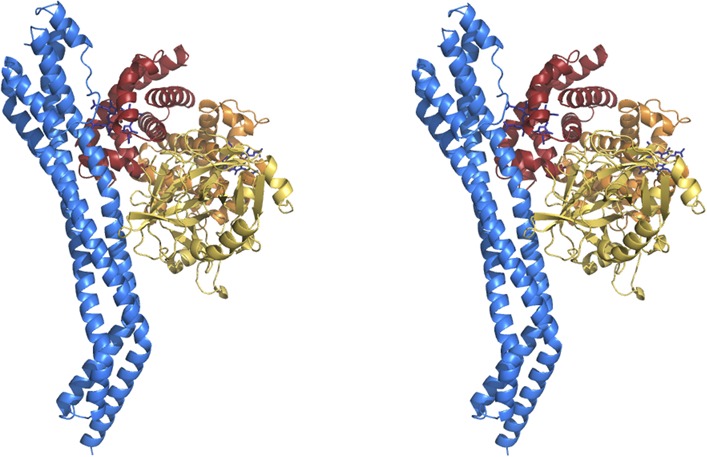
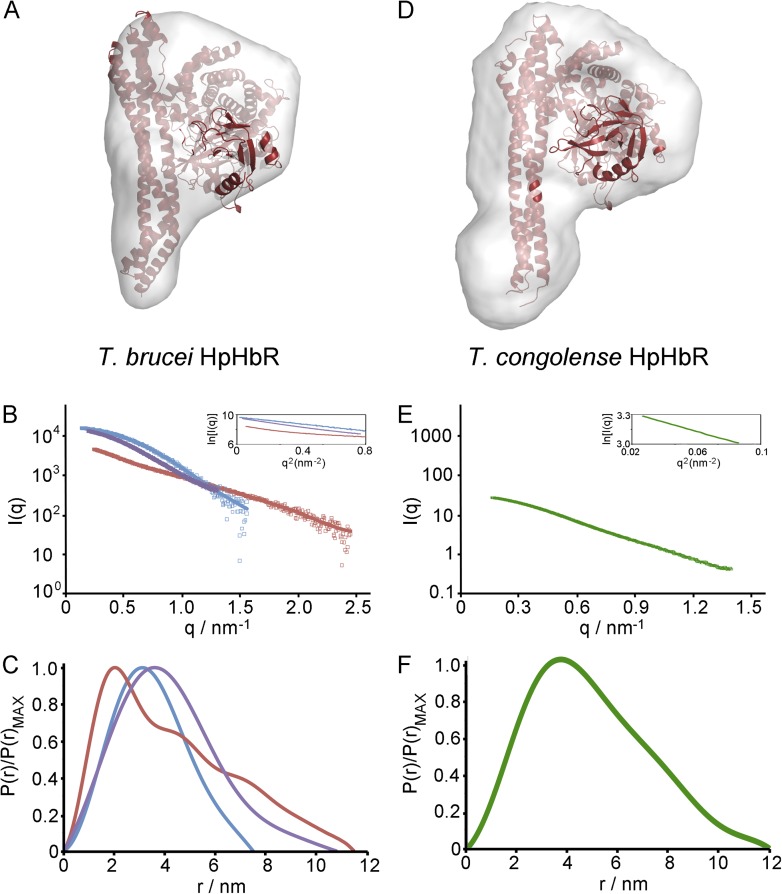
Figure 2—figure supplement 2. Small angle x-ray scattering of complexes of TcHpHbR and TbHpHbR with HpSPHb.
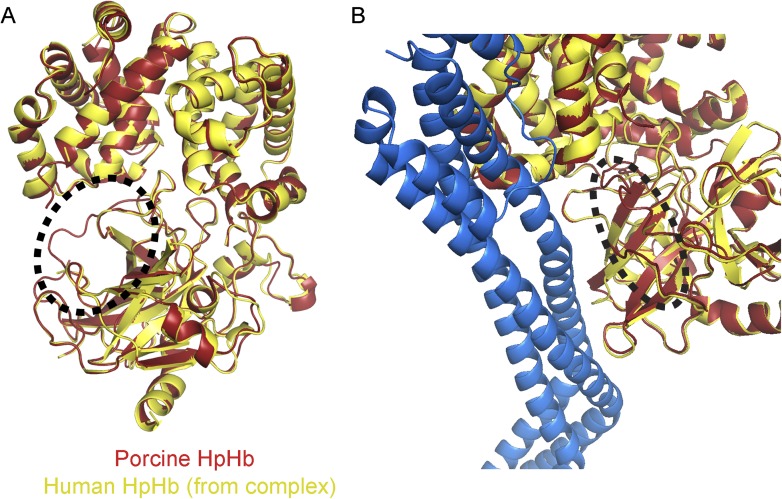
Figure 2—figure supplement 3. Clashes between TbHpHbR and a haemoglobin tetramer explain why the receptor does not bind to haemoglobin.