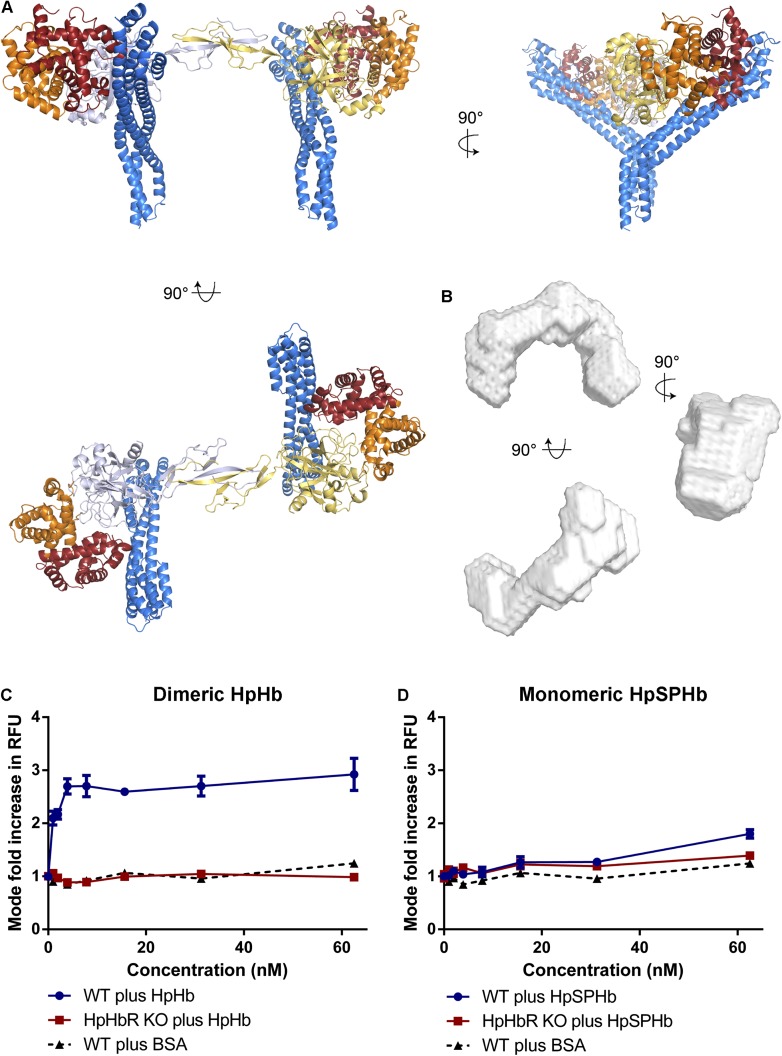
Figure 4. Simultaneous binding of two receptors to each HpHb dimer leads to more efficient uptake into trypanosomes.
(A) A model for a complex of one HpHb dimer bound to two receptors, generated by docking the structure of the TbHpHbR:HpSPHb complex onto that of porcine HpHb (Andersen et al., 2012). The receptors are organized such that two receptors, both associated with the membrane through attachment at their C-termini, can simultaneously bind to one HpHb dimer. (B) An ab initio molecular envelope derived from small angle x-ray scattering analysis of the TbHpHbR:HpHb complex supports the formation of a complex containing one HpHb dimer bound to two receptors. (C) Uptake of fluorescently labelled dimeric HpHb into live cells was monitored via flow cytometry across a range of 1–62.5 nM. Uptake saturated by 4 nM in wild-type cells whereas no uptake was observed in the HpHbR null cell line. No fluid phase uptake of labelled BSA was observed at these concentrations. (D) Uptake of fluorescently labelled monomeric HpSPHb was not readily detected until 62.5 nM, at which point uptake had not saturated. HpSPHb uptake at 62.5 nM was lost in the HpHbR null cell line. Each uptake assay was carried out in triplicate. Error bars represent standard error of the mean, n = 3.
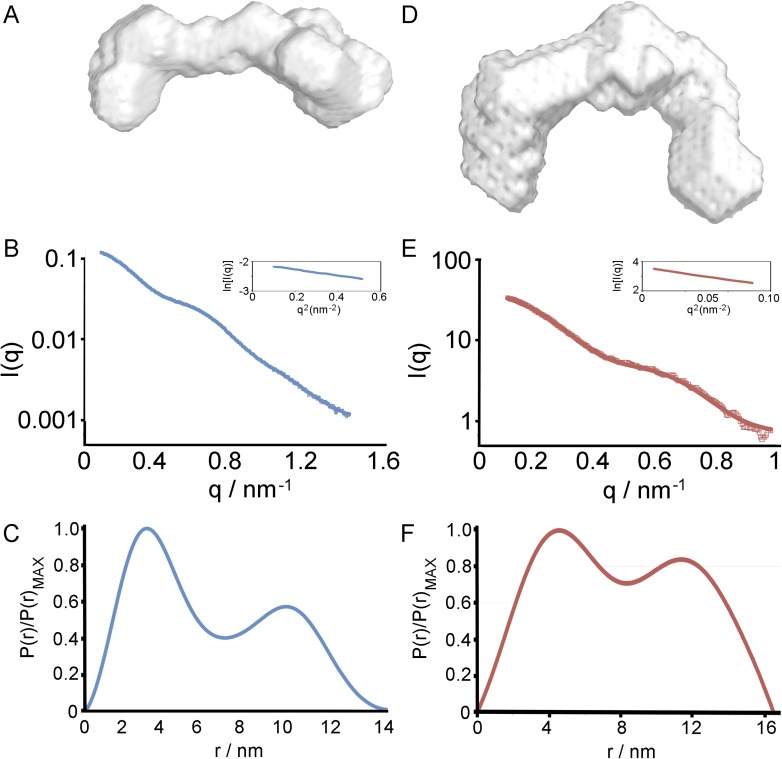
Figure 4—figure supplement 1. Small angle x-ray scattering of HpHb, alone and in complex with TbHpHbR.
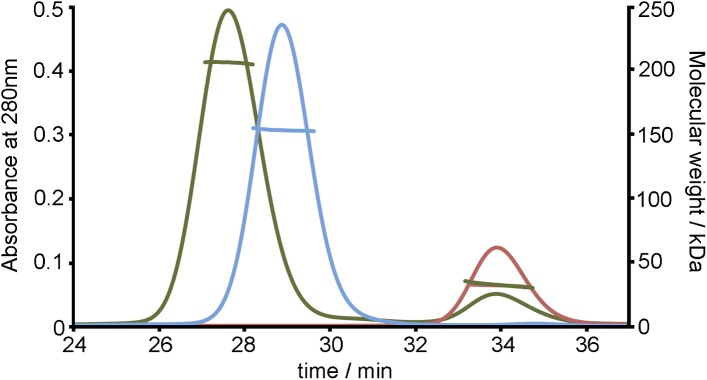
Figure 4—figure supplement 2. SEC MALLS data to assess the stoichiometry of the TbHpHbR:HpHb complex.