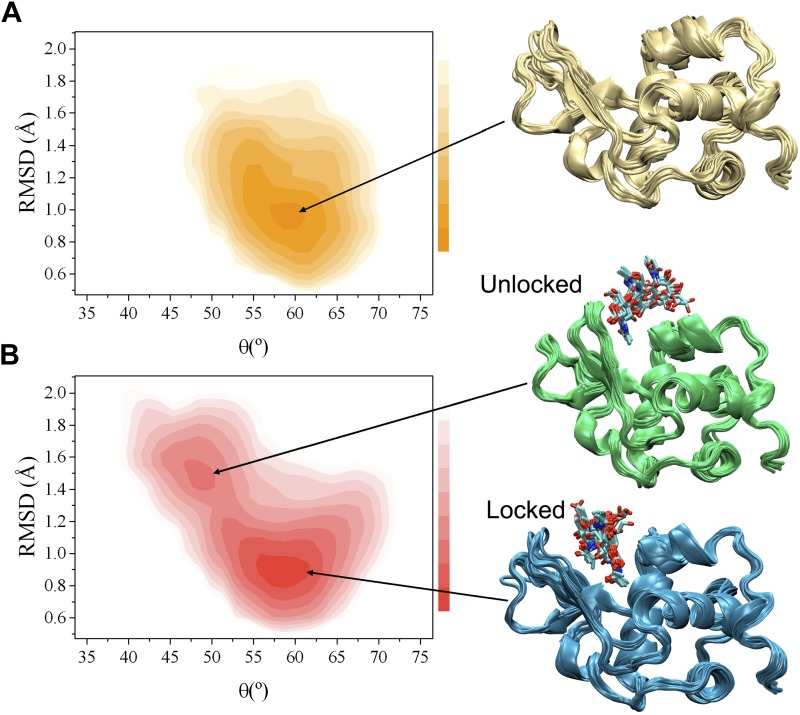
Figure 1. Comparison of the free-energy landscapes of human lysozyme in the free state (A) and in the bound state with triNAG (B).
The bound state exhibits a ground state (the ‘locked state’) and an additional local minimum with about 13% population (the ‘unlocked state’), which represents an intermediate in the release of the product of the enzymatic reaction. Free-energy landscapes are shown as function of the ‘breathing’ angle θ and of the RMSD from the X-ray structure, which was calculated on the Cα atoms by including secondary structure regions only, of a human lysozyme variant in complex with triNAG (PDB code 1BB5); free-energy landscapes were obtained as −kBTlnH(θ, RMSD), where H(θ, RMSD) is the number of times conformations with specific θ and RMSD values was sampled during the simulations (De Simone et al., 2013b).
Figure 1—figure supplement 1. Assignments of the 1H-15N HSQC spectra of the free and triNAG-bound states of human lysozyme.
Figure 1—figure supplement 2. Extracts of 1H-15N HSQC spectra showing the titration of triNAG to human lysozyme for selected residues showing significant chemical shift changes upon binding.
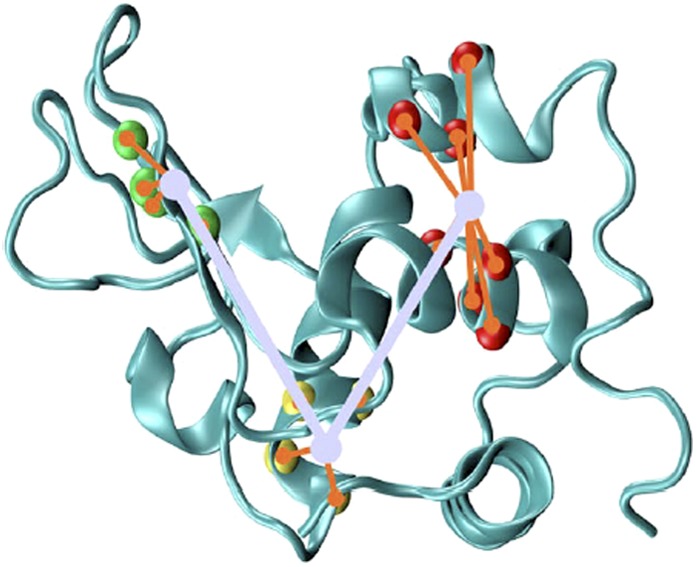
Figure 1—figure supplement 3. Illustration of the breathing angle θ of lysozyme (De Simone et al., 2013b), which accounts for the large-amplitude motion between the α-domain and β-domain of lysozyme and is computed from the centres of mass of Cα-atoms from three protein regions (De Simone et al., 2013b).